Shih-Cheng Huang
Adaptive Helpfulness-Harmlessness Alignment with Preference Vectors
Apr 27, 2025Abstract:Ensuring that large language models (LLMs) are both helpful and harmless is a critical challenge, as overly strict constraints can lead to excessive refusals, while permissive models risk generating harmful content. Existing approaches, such as reinforcement learning from human feedback (RLHF) and direct preference optimization (DPO), attempt to balance these trade-offs but suffer from performance conflicts, limited controllability, and poor extendability. To address these issues, we propose Preference Vector, a novel framework inspired by task arithmetic. Instead of optimizing multiple preferences within a single objective, we train separate models on individual preferences, extract behavior shifts as preference vectors, and dynamically merge them at test time. This modular approach enables fine-grained, user-controllable preference adjustments and facilitates seamless integration of new preferences without retraining. Experiments show that our proposed Preference Vector framework improves helpfulness without excessive conservatism, allows smooth control over preference trade-offs, and supports scalable multi-preference alignment.
CheXpert Plus: Augmenting a Large Chest X-ray Dataset with Text Radiology Reports, Patient Demographics and Additional Image Formats
Jun 03, 2024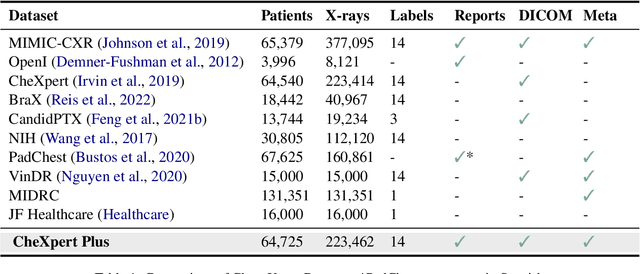

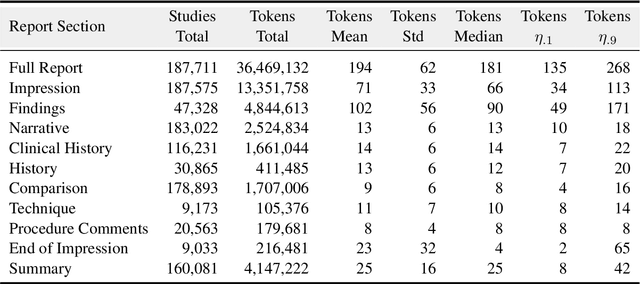
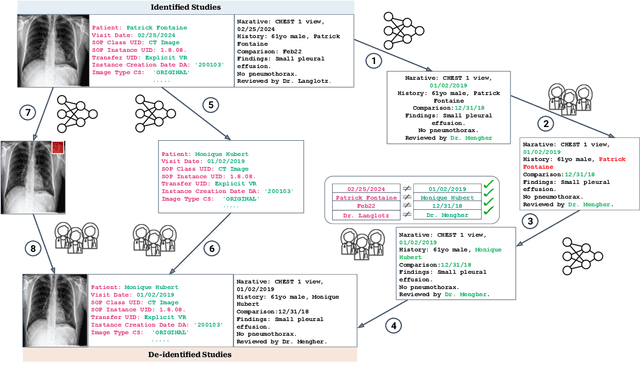
Abstract:Since the release of the original CheXpert paper five years ago, CheXpert has become one of the most widely used and cited clinical AI datasets. The emergence of vision language models has sparked an increase in demands for sharing reports linked to CheXpert images, along with a growing interest among AI fairness researchers in obtaining demographic data. To address this, CheXpert Plus serves as a new collection of radiology data sources, made publicly available to enhance the scaling, performance, robustness, and fairness of models for all subsequent machine learning tasks in the field of radiology. CheXpert Plus is the largest text dataset publicly released in radiology, with a total of 36 million text tokens, including 13 million impression tokens. To the best of our knowledge, it represents the largest text de-identification effort in radiology, with almost 1 million PHI spans anonymized. It is only the second time that a large-scale English paired dataset has been released in radiology, thereby enabling, for the first time, cross-institution training at scale. All reports are paired with high-quality images in DICOM format, along with numerous image and patient metadata covering various clinical and socio-economic groups, as well as many pathology labels and RadGraph annotations. We hope this dataset will boost research for AI models that can further assist radiologists and help improve medical care. Data is available at the following URL: https://stanfordaimi.azurewebsites.net/datasets/5158c524-d3ab-4e02-96e9-6ee9efc110a1 Models are available at the following URL: https://github.com/Stanford-AIMI/chexpert-plus
Training Small Multimodal Models to Bridge Biomedical Competency Gap: A Case Study in Radiology Imaging
Mar 20, 2024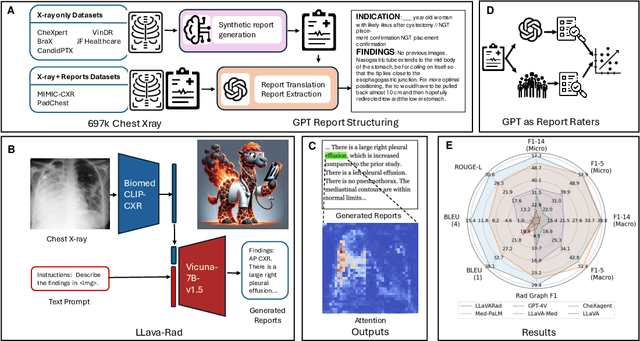
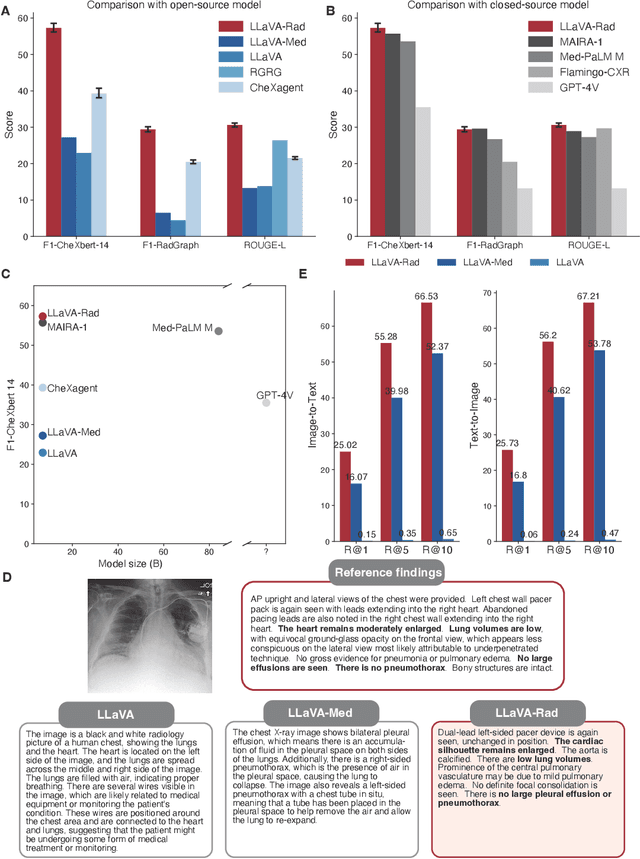
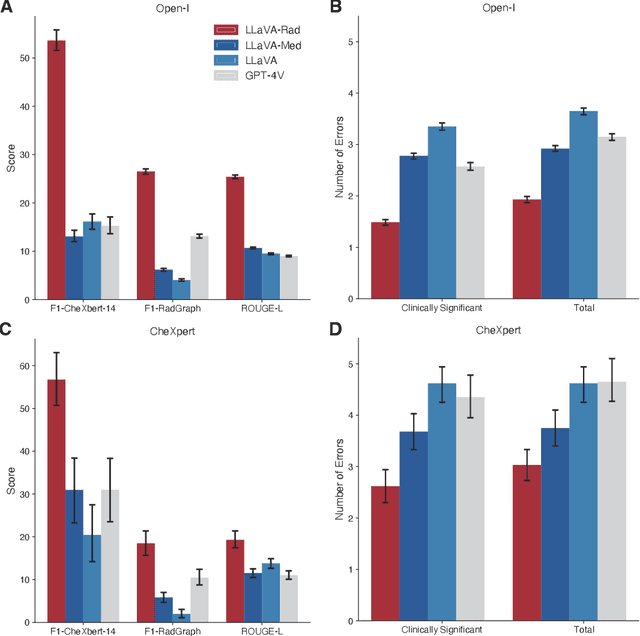
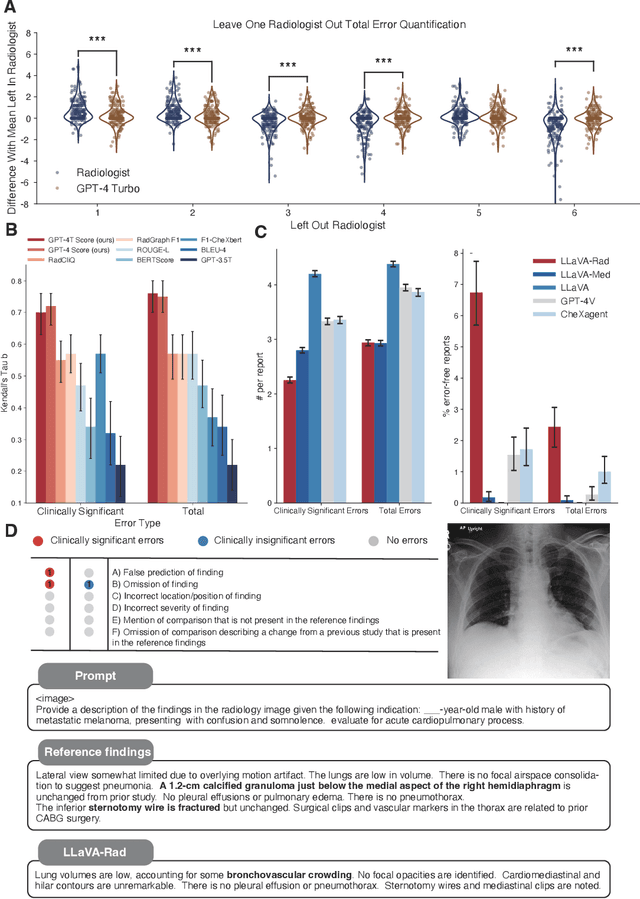
Abstract:The scaling laws and extraordinary performance of large foundation models motivate the development and utilization of such large models in biomedicine. However, despite early promising results on some biomedical benchmarks, there are still major challenges that need to be addressed before these models can be used in real-world applications. Frontier models such as GPT-4V still have major competency gaps in multimodal capabilities for biomedical applications. Moreover, pragmatic issues such as access, cost, latency, and compliance make it hard for clinicians to use privately-hosted state-of-the-art large models directly on private patient data. In this paper, we explore training open-source small multimodal models (SMMs) to bridge biomedical competency gaps for unmet clinical needs. To maximize data efficiency, we adopt a modular approach by incorporating state-of-the-art pre-trained models for image and text modalities, and focusing on training a lightweight adapter to ground each modality to the text embedding space. We conduct a comprehensive study of this approach on radiology imaging. For training, we assemble a large dataset with over 1 million image-text pairs. For evaluation, we propose a clinically driven novel approach using GPT-4 and demonstrate its parity with expert evaluation. We also study grounding qualitatively using attention. For best practice, we conduct a systematic ablation study on various choices in data engineering and multimodal training. The resulting LLaVA-Rad (7B) model attains state-of-the-art results on radiology tasks such as report generation and cross-modal retrieval, even outperforming much larger models such as GPT-4V and Med-PaLM M (84B). LLaVA-Rad is fast and can be run on a single V100 GPU in private settings, offering a promising state-of-the-art tool for real-world clinical applications.
INSPECT: A Multimodal Dataset for Pulmonary Embolism Diagnosis and Prognosis
Nov 17, 2023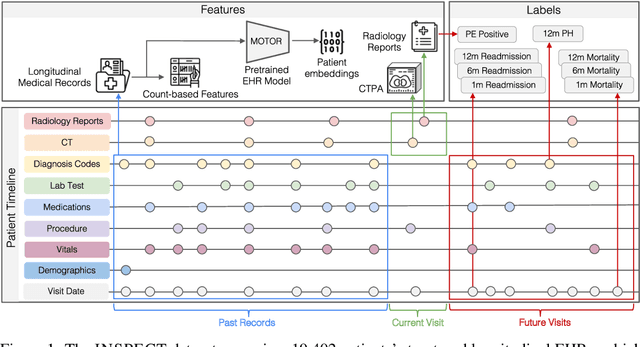
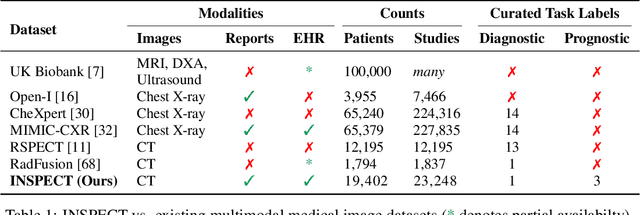
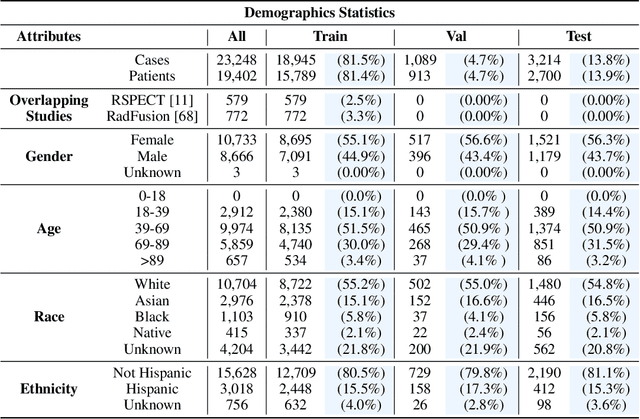
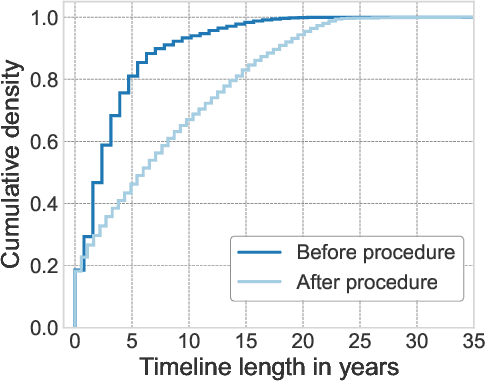
Abstract:Synthesizing information from multiple data sources plays a crucial role in the practice of modern medicine. Current applications of artificial intelligence in medicine often focus on single-modality data due to a lack of publicly available, multimodal medical datasets. To address this limitation, we introduce INSPECT, which contains de-identified longitudinal records from a large cohort of patients at risk for pulmonary embolism (PE), along with ground truth labels for multiple outcomes. INSPECT contains data from 19,402 patients, including CT images, radiology report impression sections, and structured electronic health record (EHR) data (i.e. demographics, diagnoses, procedures, vitals, and medications). Using INSPECT, we develop and release a benchmark for evaluating several baseline modeling approaches on a variety of important PE related tasks. We evaluate image-only, EHR-only, and multimodal fusion models. Trained models and the de-identified dataset are made available for non-commercial use under a data use agreement. To the best of our knowledge, INSPECT is the largest multimodal dataset integrating 3D medical imaging and EHR for reproducible methods evaluation and research.
Chat Vector: A Simple Approach to Equip LLMs With New Language Chat Capabilities
Oct 07, 2023
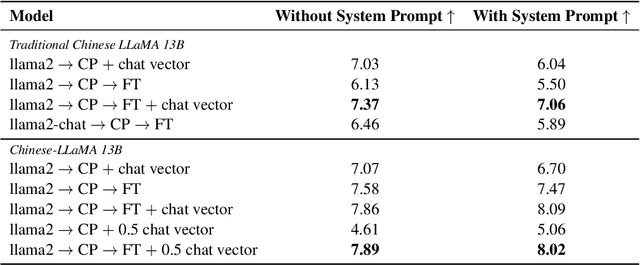

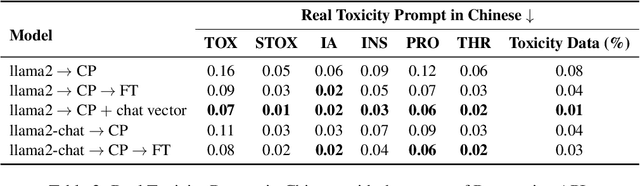
Abstract:With the advancements in conversational AI, such as ChatGPT, this paper focuses on exploring developing Large Language Models (LLMs) for non-English languages, especially emphasizing alignment with human preferences. We introduce a computationally efficient method, leveraging chat vector, to synergize pre-existing knowledge and behaviors in LLMs, restructuring the conventional training paradigm from continual pre-train -> SFT -> RLHF to continual pre-train + chat vector. Our empirical studies, primarily focused on Traditional Chinese, employ LLaMA2 as the base model and acquire the chat vector by subtracting the pre-trained weights, LLaMA2, from the weights of LLaMA2-chat. Evaluating from three distinct facets, which are toxicity, ability of instruction following, and multi-turn dialogue demonstrates the chat vector's superior efficacy in chatting. To confirm the adaptability of our approach, we extend our experiments to include models pre-trained in both Korean and Simplified Chinese, illustrating the versatility of our methodology. Overall, we present a significant solution in aligning LLMs with human preferences efficiently across various languages, accomplished by the chat vector.
LOVM: Language-Only Vision Model Selection
Jun 15, 2023Abstract:Pre-trained multi-modal vision-language models (VLMs) are becoming increasingly popular due to their exceptional performance on downstream vision applications, particularly in the few- and zero-shot settings. However, selecting the best-performing VLM for some downstream applications is non-trivial, as it is dataset and task-dependent. Meanwhile, the exhaustive evaluation of all available VLMs on a novel application is not only time and computationally demanding but also necessitates the collection of a labeled dataset for evaluation. As the number of open-source VLM variants increases, there is a need for an efficient model selection strategy that does not require access to a curated evaluation dataset. This paper proposes a novel task and benchmark for efficiently evaluating VLMs' zero-shot performance on downstream applications without access to the downstream task dataset. Specifically, we introduce a new task LOVM: Language-Only Vision Model Selection, where methods are expected to perform both model selection and performance prediction based solely on a text description of the desired downstream application. We then introduced an extensive LOVM benchmark consisting of ground-truth evaluations of 35 pre-trained VLMs and 23 datasets, where methods are expected to rank the pre-trained VLMs and predict their zero-shot performance.
BenchMD: A Benchmark for Modality-Agnostic Learning on Medical Images and Sensors
Apr 17, 2023



Abstract:Medical data poses a daunting challenge for AI algorithms: it exists in many different modalities, experiences frequent distribution shifts, and suffers from a scarcity of examples and labels. Recent advances, including transformers and self-supervised learning, promise a more universal approach that can be applied flexibly across these diverse conditions. To measure and drive progress in this direction, we present BenchMD: a benchmark that tests how modality-agnostic methods, including architectures and training techniques (e.g. self-supervised learning, ImageNet pretraining), perform on a diverse array of clinically-relevant medical tasks. BenchMD combines 19 publicly available datasets for 7 medical modalities, including 1D sensor data, 2D images, and 3D volumetric scans. Our benchmark reflects real-world data constraints by evaluating methods across a range of dataset sizes, including challenging few-shot settings that incentivize the use of pretraining. Finally, we evaluate performance on out-of-distribution data collected at different hospitals than the training data, representing naturally-occurring distribution shifts that frequently degrade the performance of medical AI models. Our baseline results demonstrate that no modality-agnostic technique achieves strong performance across all modalities, leaving ample room for improvement on the benchmark. Code is released at https://github.com/rajpurkarlab/BenchMD .
Video Pretraining Advances 3D Deep Learning on Chest CT Tasks
Apr 02, 2023Abstract:Pretraining on large natural image classification datasets such as ImageNet has aided model development on data-scarce 2D medical tasks. 3D medical tasks often have much less data than 2D medical tasks, prompting practitioners to rely on pretrained 2D models to featurize slices. However, these 2D models have been surpassed by 3D models on 3D computer vision benchmarks since they do not natively leverage cross-sectional or temporal information. In this study, we explore whether natural video pretraining for 3D models can enable higher performance on smaller datasets for 3D medical tasks. We demonstrate video pretraining improves the average performance of seven 3D models on two chest CT datasets, regardless of finetuning dataset size, and that video pretraining allows 3D models to outperform 2D baselines. Lastly, we observe that pretraining on the large-scale out-of-domain Kinetics dataset improves performance more than pretraining on a typically-sized in-domain CT dataset. Our results show consistent benefits of video pretraining across a wide array of architectures, tasks, and training dataset sizes, supporting a shift from small-scale in-domain pretraining to large-scale out-of-domain pretraining for 3D medical tasks. Our code is available at: https://github.com/rajpurkarlab/chest-ct-pretraining
Adapting Pre-trained Vision Transformers from 2D to 3D through Weight Inflation Improves Medical Image Segmentation
Feb 08, 2023Abstract:Given the prevalence of 3D medical imaging technologies such as MRI and CT that are widely used in diagnosing and treating diverse diseases, 3D segmentation is one of the fundamental tasks of medical image analysis. Recently, Transformer-based models have started to achieve state-of-the-art performances across many vision tasks, through pre-training on large-scale natural image benchmark datasets. While works on medical image analysis have also begun to explore Transformer-based models, there is currently no optimal strategy to effectively leverage pre-trained Transformers, primarily due to the difference in dimensionality between 2D natural images and 3D medical images. Existing solutions either split 3D images into 2D slices and predict each slice independently, thereby losing crucial depth-wise information, or modify the Transformer architecture to support 3D inputs without leveraging pre-trained weights. In this work, we use a simple yet effective weight inflation strategy to adapt pre-trained Transformers from 2D to 3D, retaining the benefit of both transfer learning and depth information. We further investigate the effectiveness of transfer from different pre-training sources and objectives. Our approach achieves state-of-the-art performances across a broad range of 3D medical image datasets, and can become a standard strategy easily utilized by all work on Transformer-based models for 3D medical images, to maximize performance.
Diagnosing and Rectifying Vision Models using Language
Feb 08, 2023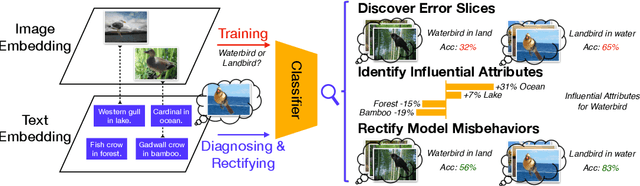
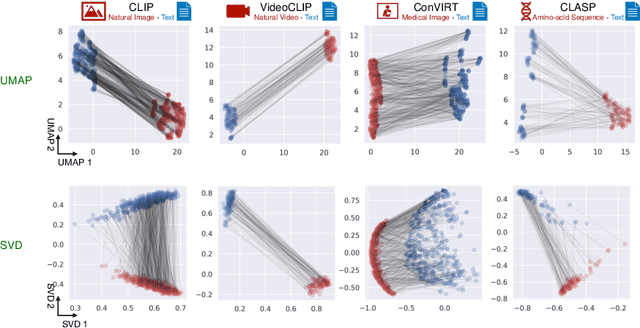
Abstract:Recent multi-modal contrastive learning models have demonstrated the ability to learn an embedding space suitable for building strong vision classifiers, by leveraging the rich information in large-scale image-caption datasets. Our work highlights a distinct advantage of this multi-modal embedding space: the ability to diagnose vision classifiers through natural language. The traditional process of diagnosing model behaviors in deployment settings involves labor-intensive data acquisition and annotation. Our proposed method can discover high-error data slices, identify influential attributes and further rectify undesirable model behaviors, without requiring any visual data. Through a combination of theoretical explanation and empirical verification, we present conditions under which classifiers trained on embeddings from one modality can be equivalently applied to embeddings from another modality. On a range of image datasets with known error slices, we demonstrate that our method can effectively identify the error slices and influential attributes, and can further use language to rectify failure modes of the classifier.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge