Yanbo Xu
Scaling medical imaging report generation with multimodal reinforcement learning
Jan 23, 2026Abstract:Frontier models have demonstrated remarkable capabilities in understanding and reasoning with natural-language text, but they still exhibit major competency gaps in multimodal understanding and reasoning especially in high-value verticals such as biomedicine. Medical imaging report generation is a prominent example. Supervised fine-tuning can substantially improve performance, but they are prone to overfitting to superficial boilerplate patterns. In this paper, we introduce Universal Report Generation (UniRG) as a general framework for medical imaging report generation. By leveraging reinforcement learning as a unifying mechanism to directly optimize for evaluation metrics designed for end applications, UniRG can significantly improve upon supervised fine-tuning and attain durable generalization across diverse institutions and clinical practices. We trained UniRG-CXR on publicly available chest X-ray (CXR) data and conducted a thorough evaluation in CXR report generation with rigorous evaluation scenarios. On the authoritative ReXrank benchmark, UniRG-CXR sets new overall SOTA, outperforming prior state of the art by a wide margin.
Universal Abstraction: Harnessing Frontier Models to Structure Real-World Data at Scale
Feb 02, 2025



Abstract:The vast majority of real-world patient information resides in unstructured clinical text, and the process of medical abstraction seeks to extract and normalize structured information from this unstructured input. However, traditional medical abstraction methods can require significant manual efforts that can include crafting rules or annotating training labels, limiting scalability. In this paper, we propose UniMedAbstractor (UMA), a zero-shot medical abstraction framework leveraging Large Language Models (LLMs) through a modular and customizable prompt template. We refer to our approach as universal abstraction as it can quickly scale to new attributes through its universal prompt template without curating attribute-specific training labels or rules. We evaluate UMA for oncology applications, focusing on fifteen key attributes representing the cancer patient journey, from short-context attributes (e.g., performance status, treatment) to complex long-context attributes requiring longitudinal reasoning (e.g., tumor site, histology, TNM staging). Experiments on real-world data show UMA's strong performance and generalizability. Compared to supervised and heuristic baselines, UMA with GPT-4o achieves on average an absolute 2-point F1/accuracy improvement for both short-context and long-context attribute abstraction. For pathologic T staging, UMA even outperforms the supervised model by 20 points in accuracy.
Diverse Score Distillation
Dec 09, 2024



Abstract:Score distillation of 2D diffusion models has proven to be a powerful mechanism to guide 3D optimization, for example enabling text-based 3D generation or single-view reconstruction. A common limitation of existing score distillation formulations, however, is that the outputs of the (mode-seeking) optimization are limited in diversity despite the underlying diffusion model being capable of generating diverse samples. In this work, inspired by the sampling process in denoising diffusion, we propose a score formulation that guides the optimization to follow generation paths defined by random initial seeds, thus ensuring diversity. We then present an approximation to adopt this formulation for scenarios where the optimization may not precisely follow the generation paths (e.g. a 3D representation whose renderings evolve in a co-dependent manner). We showcase the applications of our `Diverse Score Distillation' (DSD) formulation across tasks such as 2D optimization, text-based 3D inference, and single-view reconstruction. We also empirically validate DSD against prior score distillation formulations and show that it significantly improves sample diversity while preserving fidelity.
Training Small Multimodal Models to Bridge Biomedical Competency Gap: A Case Study in Radiology Imaging
Mar 20, 2024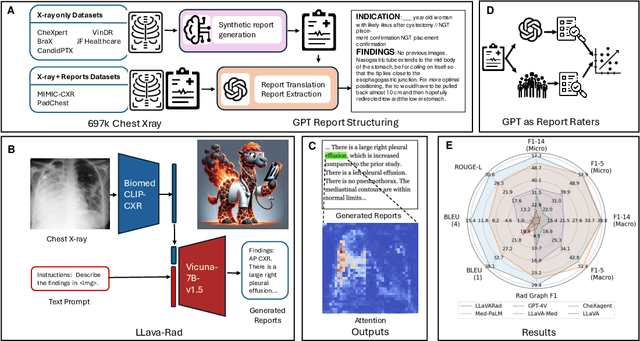
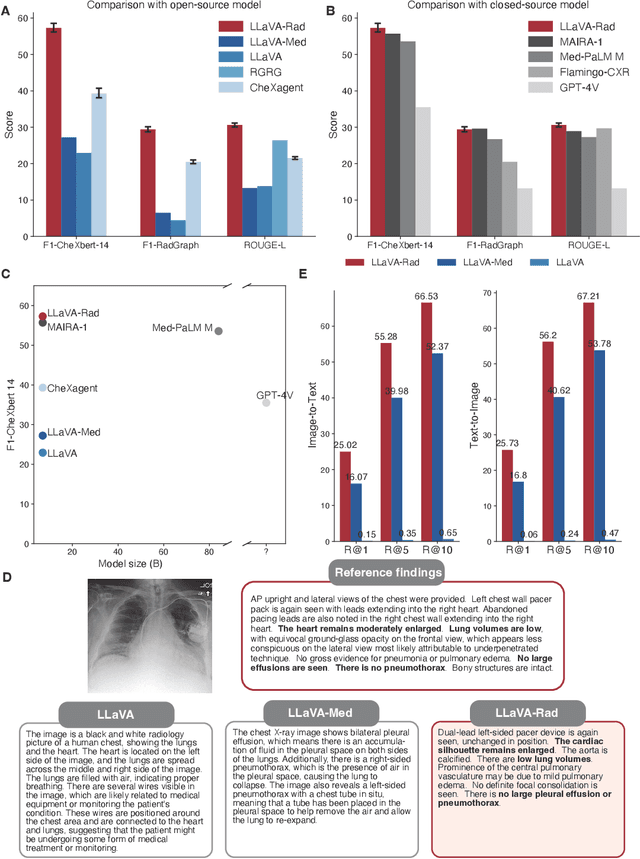
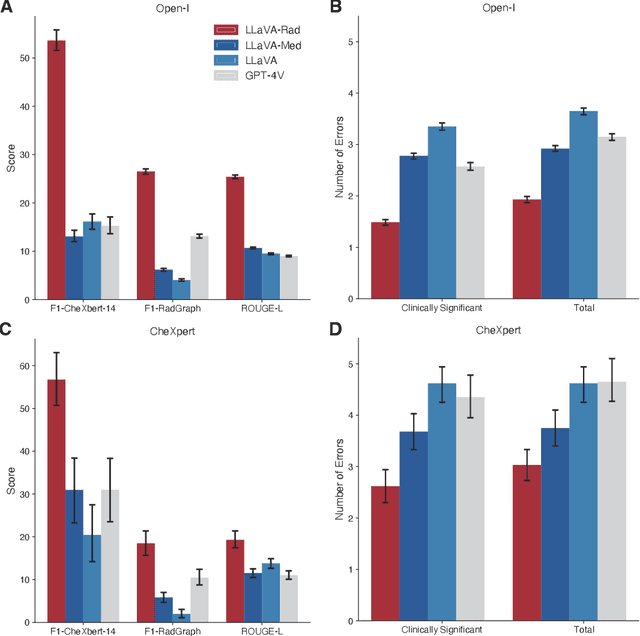
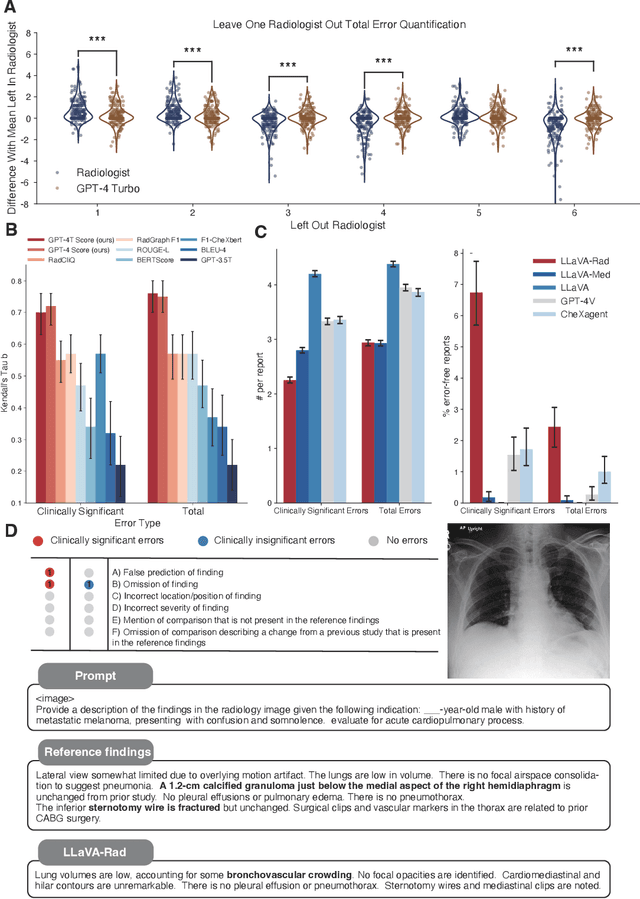
Abstract:The scaling laws and extraordinary performance of large foundation models motivate the development and utilization of such large models in biomedicine. However, despite early promising results on some biomedical benchmarks, there are still major challenges that need to be addressed before these models can be used in real-world applications. Frontier models such as GPT-4V still have major competency gaps in multimodal capabilities for biomedical applications. Moreover, pragmatic issues such as access, cost, latency, and compliance make it hard for clinicians to use privately-hosted state-of-the-art large models directly on private patient data. In this paper, we explore training open-source small multimodal models (SMMs) to bridge biomedical competency gaps for unmet clinical needs. To maximize data efficiency, we adopt a modular approach by incorporating state-of-the-art pre-trained models for image and text modalities, and focusing on training a lightweight adapter to ground each modality to the text embedding space. We conduct a comprehensive study of this approach on radiology imaging. For training, we assemble a large dataset with over 1 million image-text pairs. For evaluation, we propose a clinically driven novel approach using GPT-4 and demonstrate its parity with expert evaluation. We also study grounding qualitatively using attention. For best practice, we conduct a systematic ablation study on various choices in data engineering and multimodal training. The resulting LLaVA-Rad (7B) model attains state-of-the-art results on radiology tasks such as report generation and cross-modal retrieval, even outperforming much larger models such as GPT-4V and Med-PaLM M (84B). LLaVA-Rad is fast and can be run on a single V100 GPU in private settings, offering a promising state-of-the-art tool for real-world clinical applications.
Foundation Models for Biomedical Image Segmentation: A Survey
Jan 15, 2024Abstract:Recent advancements in biomedical image analysis have been significantly driven by the Segment Anything Model (SAM). This transformative technology, originally developed for general-purpose computer vision, has found rapid application in medical image processing. Within the last year, marked by over 100 publications, SAM has demonstrated its prowess in zero-shot learning adaptations for medical imaging. The fundamental premise of SAM lies in its capability to segment or identify objects in images without prior knowledge of the object type or imaging modality. This approach aligns well with tasks achievable by the human visual system, though its application in non-biological vision contexts remains more theoretically challenging. A notable feature of SAM is its ability to adjust segmentation according to a specified resolution scale or area of interest, akin to semantic priming. This adaptability has spurred a wave of creativity and innovation in applying SAM to medical imaging. Our review focuses on the period from April 1, 2023, to September 30, 2023, a critical first six months post-initial publication. We examine the adaptations and integrations of SAM necessary to address longstanding clinical challenges, particularly in the context of 33 open datasets covered in our analysis. While SAM approaches or achieves state-of-the-art performance in numerous applications, it falls short in certain areas, such as segmentation of the carotid artery, adrenal glands, optic nerve, and mandible bone. Our survey delves into the innovative techniques where SAM's foundational approach excels and explores the core concepts in translating and applying these models effectively in diverse medical imaging scenarios.
SMURF-THP: Score Matching-based UnceRtainty quantiFication for Transformer Hawkes Process
Oct 25, 2023
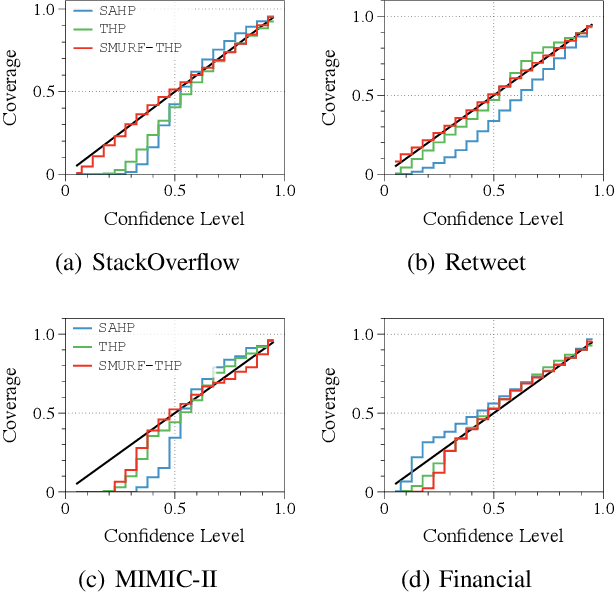
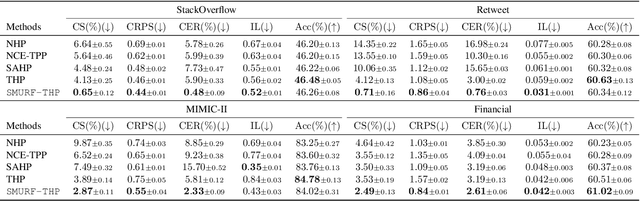
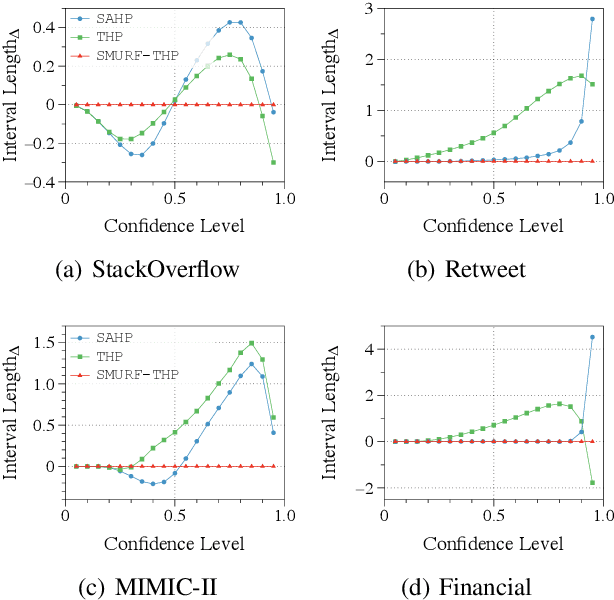
Abstract:Transformer Hawkes process models have shown to be successful in modeling event sequence data. However, most of the existing training methods rely on maximizing the likelihood of event sequences, which involves calculating some intractable integral. Moreover, the existing methods fail to provide uncertainty quantification for model predictions, e.g., confidence intervals for the predicted event's arrival time. To address these issues, we propose SMURF-THP, a score-based method for learning Transformer Hawkes process and quantifying prediction uncertainty. Specifically, SMURF-THP learns the score function of events' arrival time based on a score-matching objective that avoids the intractable computation. With such a learned score function, we can sample arrival time of events from the predictive distribution. This naturally allows for the quantification of uncertainty by computing confidence intervals over the generated samples. We conduct extensive experiments in both event type prediction and uncertainty quantification of arrival time. In all the experiments, SMURF-THP outperforms existing likelihood-based methods in confidence calibration while exhibiting comparable prediction accuracy.
FDNeRF: Semantics-Driven Face Reconstruction, Prompt Editing and Relighting with Diffusion Models
Jun 01, 2023Abstract:The ability to create high-quality 3D faces from a single image has become increasingly important with wide applications in video conferencing, AR/VR, and advanced video editing in movie industries. In this paper, we propose Face Diffusion NeRF (FDNeRF), a new generative method to reconstruct high-quality Face NeRFs from single images, complete with semantic editing and relighting capabilities. FDNeRF utilizes high-resolution 3D GAN inversion and expertly trained 2D latent-diffusion model, allowing users to manipulate and construct Face NeRFs in zero-shot learning without the need for explicit 3D data. With carefully designed illumination and identity preserving loss, as well as multi-modal pre-training, FD-NeRF offers users unparalleled control over the editing process enabling them to create and edit face NeRFs using just single-view images, text prompts, and explicit target lighting. The advanced features of FDNeRF have been designed to produce more impressive results than existing 2D editing approaches that rely on 2D segmentation maps for editable attributes. Experiments show that our FDNeRF achieves exceptionally realistic results and unprecedented flexibility in editing compared with state-of-the-art 3D face reconstruction and editing methods. Our code will be available at https://github.com/BillyXYB/FDNeRF.
Large-Scale Domain-Specific Pretraining for Biomedical Vision-Language Processing
Mar 02, 2023Abstract:Contrastive pretraining on parallel image-text data has attained great success in vision-language processing (VLP), as exemplified by CLIP and related methods. However, prior explorations tend to focus on general domains in the web. Biomedical images and text are rather different, but publicly available datasets are small and skew toward chest X-ray, thus severely limiting progress. In this paper, we conducted by far the largest study on biomedical VLP, using 15 million figure-caption pairs extracted from biomedical research articles in PubMed Central. Our dataset (PMC-15M) is two orders of magnitude larger than existing biomedical image-text datasets such as MIMIC-CXR, and spans a diverse range of biomedical images. The standard CLIP method is suboptimal for the biomedical domain. We propose BiomedCLIP with domain-specific adaptations tailored to biomedical VLP. We conducted extensive experiments and ablation studies on standard biomedical imaging tasks from retrieval to classification to visual question-answering (VQA). BiomedCLIP established new state of the art in a wide range of standard datasets, substantially outperformed prior VLP approaches. Surprisingly, BiomedCLIP even outperformed radiology-specific state-of-the-art models such as BioViL on radiology-specific tasks such as RSNA pneumonia detection, thus highlighting the utility in large-scale pretraining across all biomedical image types. We will release our models at https://aka.ms/biomedclip to facilitate future research in biomedical VLP.
UnfoldML: Cost-Aware and Uncertainty-Based Dynamic 2D Prediction for Multi-Stage Classification
Oct 28, 2022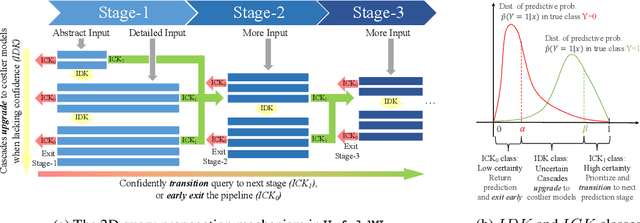

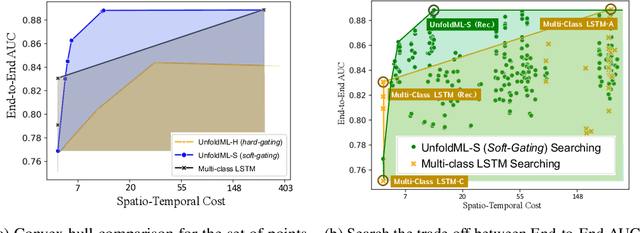
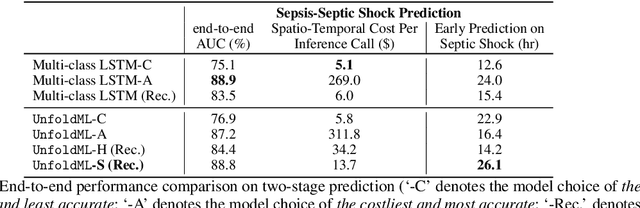
Abstract:Machine Learning (ML) research has focused on maximizing the accuracy of predictive tasks. ML models, however, are increasingly more complex, resource intensive, and costlier to deploy in resource-constrained environments. These issues are exacerbated for prediction tasks with sequential classification on progressively transitioned stages with ''happens-before'' relation between them.We argue that it is possible to ''unfold'' a monolithic single multi-class classifier, typically trained for all stages using all data, into a series of single-stage classifiers. Each single-stage classifier can be cascaded gradually from cheaper to more expensive binary classifiers that are trained using only the necessary data modalities or features required for that stage. UnfoldML is a cost-aware and uncertainty-based dynamic 2D prediction pipeline for multi-stage classification that enables (1) navigation of the accuracy/cost tradeoff space, (2) reducing the spatio-temporal cost of inference by orders of magnitude, and (3) early prediction on proceeding stages. UnfoldML achieves orders of magnitude better cost in clinical settings, while detecting multi-stage disease development in real time. It achieves within 0.1% accuracy from the highest-performing multi-class baseline, while saving close to 20X on spatio-temporal cost of inference and earlier (3.5hrs) disease onset prediction. We also show that UnfoldML generalizes to image classification, where it can predict different level of labels (from coarse to fine) given different level of abstractions of a image, saving close to 5X cost with as little as 0.4% accuracy reduction.
TransEditor: Transformer-Based Dual-Space GAN for Highly Controllable Facial Editing
Mar 31, 2022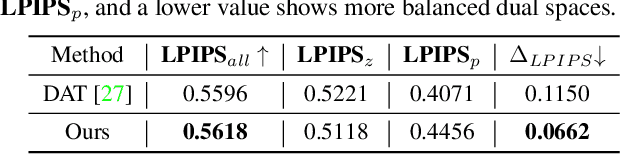
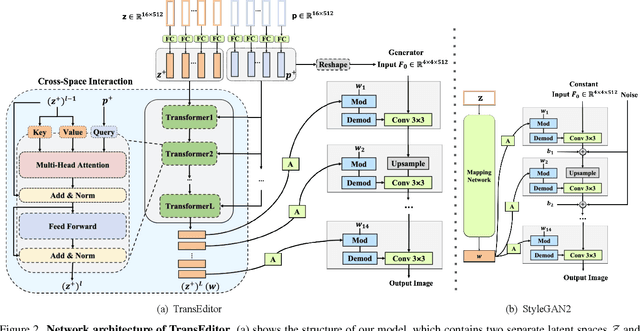
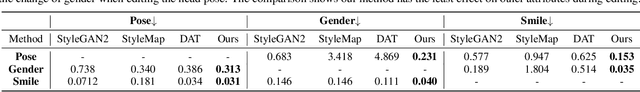
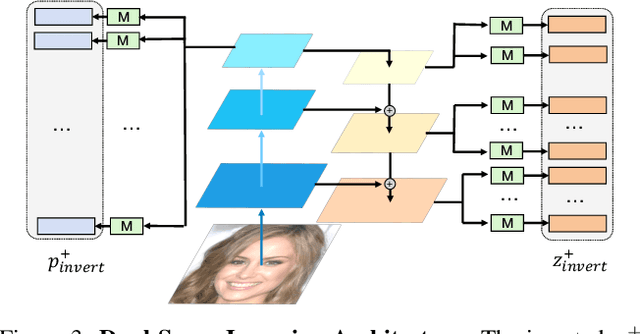
Abstract:Recent advances like StyleGAN have promoted the growth of controllable facial editing. To address its core challenge of attribute decoupling in a single latent space, attempts have been made to adopt dual-space GAN for better disentanglement of style and content representations. Nonetheless, these methods are still incompetent to obtain plausible editing results with high controllability, especially for complicated attributes. In this study, we highlight the importance of interaction in a dual-space GAN for more controllable editing. We propose TransEditor, a novel Transformer-based framework to enhance such interaction. Besides, we develop a new dual-space editing and inversion strategy to provide additional editing flexibility. Extensive experiments demonstrate the superiority of the proposed framework in image quality and editing capability, suggesting the effectiveness of TransEditor for highly controllable facial editing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge