Rajesh Rao
Large-Scale Domain-Specific Pretraining for Biomedical Vision-Language Processing
Mar 02, 2023Abstract:Contrastive pretraining on parallel image-text data has attained great success in vision-language processing (VLP), as exemplified by CLIP and related methods. However, prior explorations tend to focus on general domains in the web. Biomedical images and text are rather different, but publicly available datasets are small and skew toward chest X-ray, thus severely limiting progress. In this paper, we conducted by far the largest study on biomedical VLP, using 15 million figure-caption pairs extracted from biomedical research articles in PubMed Central. Our dataset (PMC-15M) is two orders of magnitude larger than existing biomedical image-text datasets such as MIMIC-CXR, and spans a diverse range of biomedical images. The standard CLIP method is suboptimal for the biomedical domain. We propose BiomedCLIP with domain-specific adaptations tailored to biomedical VLP. We conducted extensive experiments and ablation studies on standard biomedical imaging tasks from retrieval to classification to visual question-answering (VQA). BiomedCLIP established new state of the art in a wide range of standard datasets, substantially outperformed prior VLP approaches. Surprisingly, BiomedCLIP even outperformed radiology-specific state-of-the-art models such as BioViL on radiology-specific tasks such as RSNA pneumonia detection, thus highlighting the utility in large-scale pretraining across all biomedical image types. We will release our models at https://aka.ms/biomedclip to facilitate future research in biomedical VLP.
Towards Structuring Real-World Data at Scale: Deep Learning for Extracting Key Oncology Information from Clinical Text with Patient-Level Supervision
Mar 20, 2022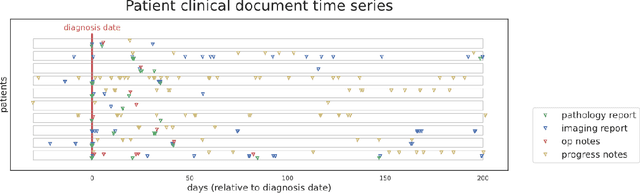
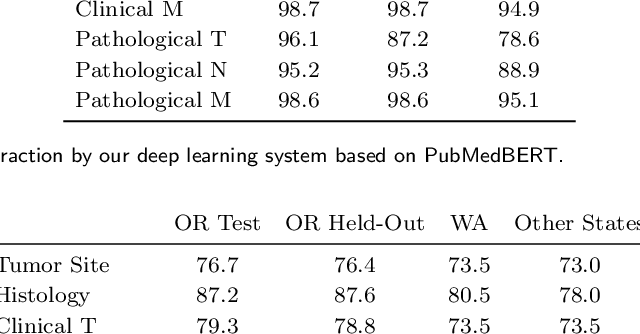
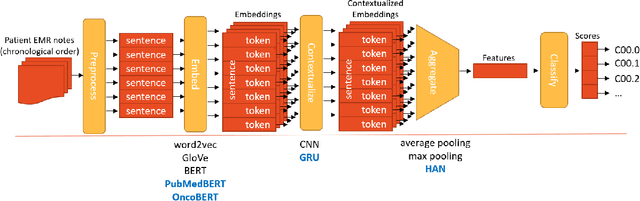
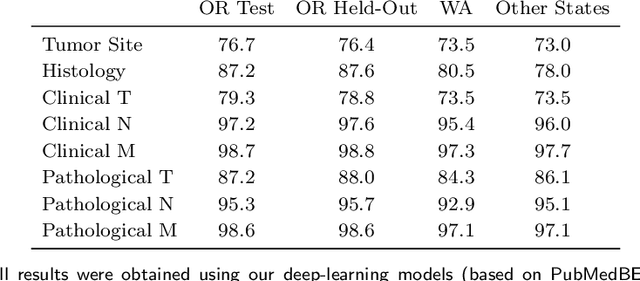
Abstract:Objective: The majority of detailed patient information in real-world data (RWD) is only consistently available in free-text clinical documents. Manual curation is expensive and time-consuming. Developing natural language processing (NLP) methods for structuring RWD is thus essential for scaling real-world evidence generation. Materials and Methods: Traditional rule-based systems are vulnerable to the prevalent linguistic variations and ambiguities in clinical text, and prior applications of machine-learning methods typically require sentence-level or report-level labeled examples that are hard to produce at scale. We propose leveraging patient-level supervision from medical registries, which are often readily available and capture key patient information, for general RWD applications. To combat the lack of sentence-level or report-level annotations, we explore advanced deep-learning methods by combining domain-specific pretraining, recurrent neural networks, and hierarchical attention. Results: We conduct an extensive study on 135,107 patients from the cancer registry of a large integrated delivery network (IDN) comprising healthcare systems in five western US states. Our deep learning methods attain test AUROC of 94-99% for key tumor attributes and comparable performance on held-out data from separate health systems and states. Discussion and Conclusion: Ablation results demonstrate clear superiority of these advanced deep-learning methods over prior approaches. Error analysis shows that our NLP system sometimes even corrects errors in registrar labels. We also conduct a preliminary investigation in accelerating registry curation and general RWD structuring via assisted curation for over 1.2 million cancer patients in this healthcare network.
Time-varying Autoregression with Low Rank Tensors
May 21, 2019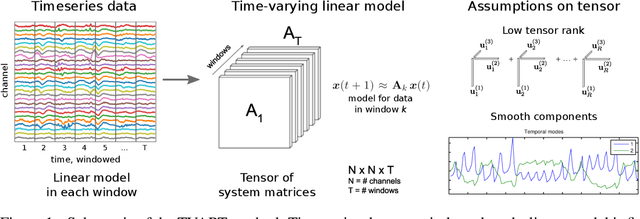
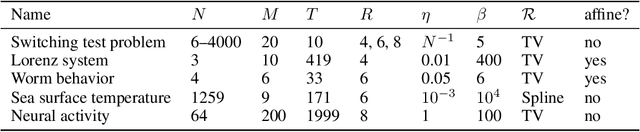
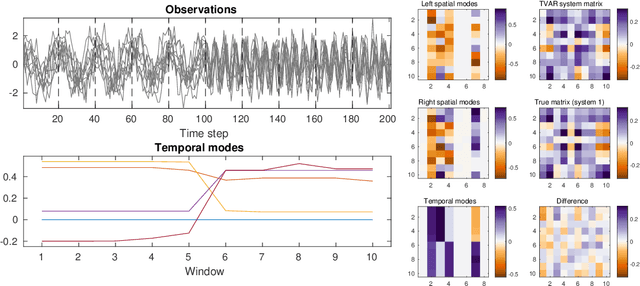
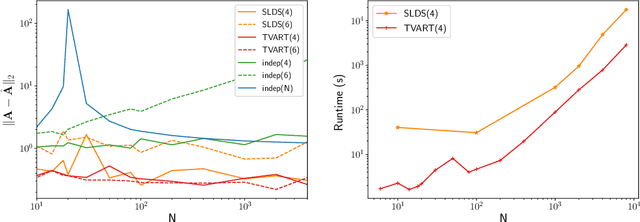
Abstract:We present a windowed technique to learn parsimonious time-varying autoregressive models from multivariate timeseries. This unsupervised method uncovers spatiotemporal structure in data via non-smooth and non-convex optimization. In each time window, we assume the data follow a linear model parameterized by a potentially different system matrix, and we model this stack of system matrices as a low rank tensor. Because of its structure, the model is scalable to high-dimensional data and can easily incorporate priors such as smoothness over time. We find the components of the tensor using alternating minimization and prove that any stationary point of this algorithm is a local minimum. In a test case, our method identifies the true rank of a switching linear system in the presence of noise. We illustrate our model's utility and superior scalability over extant methods when applied to several synthetic and real examples, including a nonlinear dynamical system, worm behavior, sea surface temperature, and monkey brain recordings.
AJILE Movement Prediction: Multimodal Deep Learning for Natural Human Neural Recordings and Video
Mar 01, 2018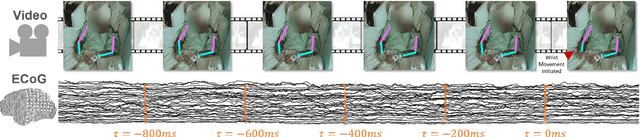
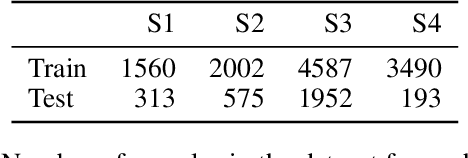
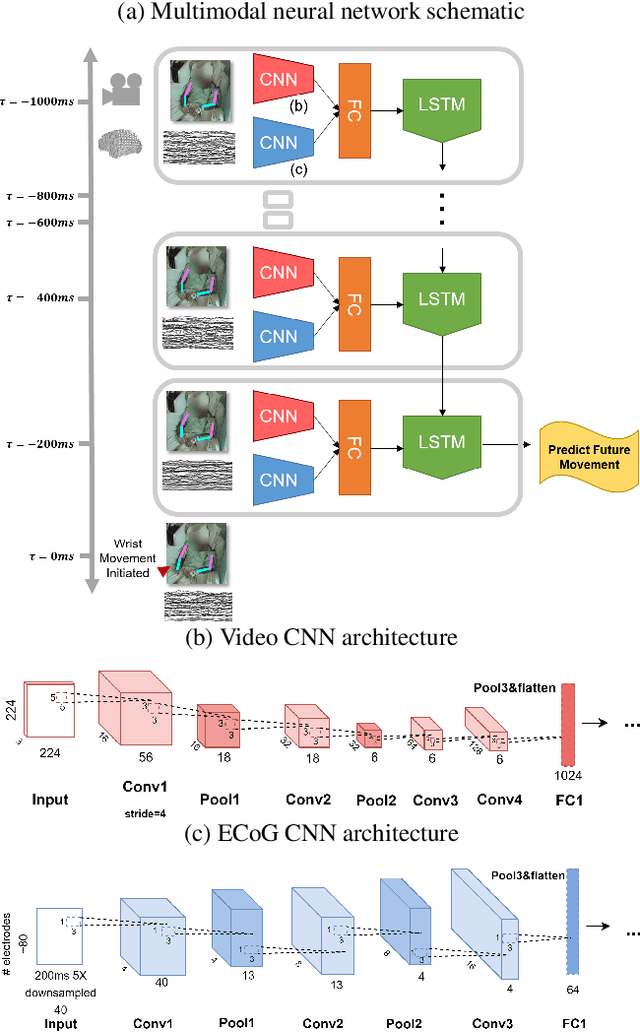
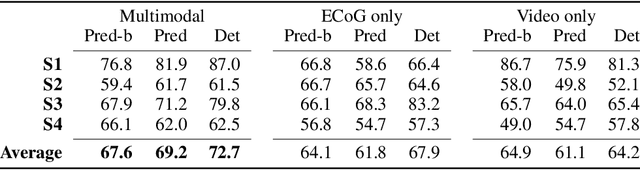
Abstract:Developing useful interfaces between brains and machines is a grand challenge of neuroengineering. An effective interface has the capacity to not only interpret neural signals, but predict the intentions of the human to perform an action in the near future; prediction is made even more challenging outside well-controlled laboratory experiments. This paper describes our approach to detect and to predict natural human arm movements in the future, a key challenge in brain computer interfacing that has never before been attempted. We introduce the novel Annotated Joints in Long-term ECoG (AJILE) dataset; AJILE includes automatically annotated poses of 7 upper body joints for four human subjects over 670 total hours (more than 72 million frames), along with the corresponding simultaneously acquired intracranial neural recordings. The size and scope of AJILE greatly exceeds all previous datasets with movements and electrocorticography (ECoG), making it possible to take a deep learning approach to movement prediction. We propose a multimodal model that combines deep convolutional neural networks (CNN) with long short-term memory (LSTM) blocks, leveraging both ECoG and video modalities. We demonstrate that our models are able to detect movements and predict future movements up to 800 msec before movement initiation. Further, our multimodal movement prediction models exhibit resilience to simulated ablation of input neural signals. We believe a multimodal approach to natural neural decoding that takes context into account is critical in advancing bioelectronic technologies and human neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge