Roshanthi Weerasinghe
Universal Abstraction: Harnessing Frontier Models to Structure Real-World Data at Scale
Feb 02, 2025



Abstract:The vast majority of real-world patient information resides in unstructured clinical text, and the process of medical abstraction seeks to extract and normalize structured information from this unstructured input. However, traditional medical abstraction methods can require significant manual efforts that can include crafting rules or annotating training labels, limiting scalability. In this paper, we propose UniMedAbstractor (UMA), a zero-shot medical abstraction framework leveraging Large Language Models (LLMs) through a modular and customizable prompt template. We refer to our approach as universal abstraction as it can quickly scale to new attributes through its universal prompt template without curating attribute-specific training labels or rules. We evaluate UMA for oncology applications, focusing on fifteen key attributes representing the cancer patient journey, from short-context attributes (e.g., performance status, treatment) to complex long-context attributes requiring longitudinal reasoning (e.g., tumor site, histology, TNM staging). Experiments on real-world data show UMA's strong performance and generalizability. Compared to supervised and heuristic baselines, UMA with GPT-4o achieves on average an absolute 2-point F1/accuracy improvement for both short-context and long-context attribute abstraction. For pathologic T staging, UMA even outperforms the supervised model by 20 points in accuracy.
TRIALSCOPE: A Unifying Causal Framework for Scaling Real-World Evidence Generation with Biomedical Language Models
Nov 06, 2023Abstract:The rapid digitization of real-world data offers an unprecedented opportunity for optimizing healthcare delivery and accelerating biomedical discovery. In practice, however, such data is most abundantly available in unstructured forms, such as clinical notes in electronic medical records (EMRs), and it is generally plagued by confounders. In this paper, we present TRIALSCOPE, a unifying framework for distilling real-world evidence from population-level observational data. TRIALSCOPE leverages biomedical language models to structure clinical text at scale, employs advanced probabilistic modeling for denoising and imputation, and incorporates state-of-the-art causal inference techniques to combat common confounders. Using clinical trial specification as generic representation, TRIALSCOPE provides a turn-key solution to generate and reason with clinical hypotheses using observational data. In extensive experiments and analyses on a large-scale real-world dataset with over one million cancer patients from a large US healthcare network, we show that TRIALSCOPE can produce high-quality structuring of real-world data and generates comparable results to marquee cancer trials. In addition to facilitating in-silicon clinical trial design and optimization, TRIALSCOPE may be used to empower synthetic controls, pragmatic trials, post-market surveillance, as well as support fine-grained patient-like-me reasoning in precision diagnosis and treatment.
Scaling Clinical Trial Matching Using Large Language Models: A Case Study in Oncology
Aug 18, 2023Abstract:Clinical trial matching is a key process in health delivery and discovery. In practice, it is plagued by overwhelming unstructured data and unscalable manual processing. In this paper, we conduct a systematic study on scaling clinical trial matching using large language models (LLMs), with oncology as the focus area. Our study is grounded in a clinical trial matching system currently in test deployment at a large U.S. health network. Initial findings are promising: out of box, cutting-edge LLMs, such as GPT-4, can already structure elaborate eligibility criteria of clinical trials and extract complex matching logic (e.g., nested AND/OR/NOT). While still far from perfect, LLMs substantially outperform prior strong baselines and may serve as a preliminary solution to help triage patient-trial candidates with humans in the loop. Our study also reveals a few significant growth areas for applying LLMs to end-to-end clinical trial matching, such as context limitation and accuracy, especially in structuring patient information from longitudinal medical records.
Towards Structuring Real-World Data at Scale: Deep Learning for Extracting Key Oncology Information from Clinical Text with Patient-Level Supervision
Mar 20, 2022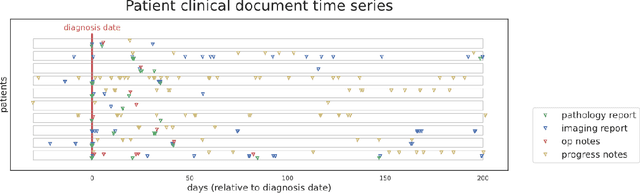
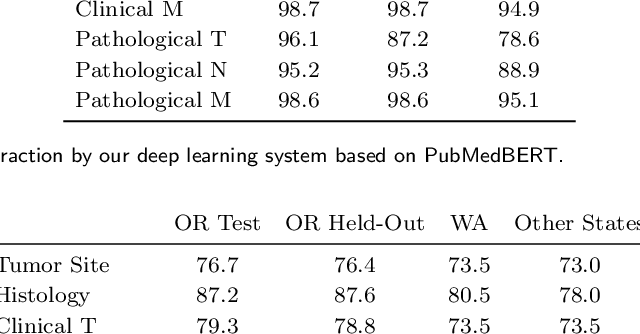
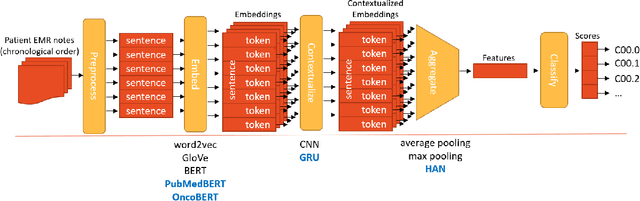
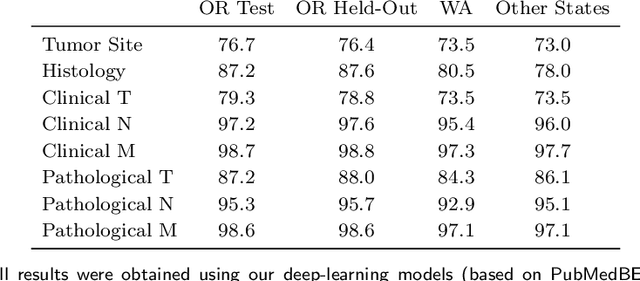
Abstract:Objective: The majority of detailed patient information in real-world data (RWD) is only consistently available in free-text clinical documents. Manual curation is expensive and time-consuming. Developing natural language processing (NLP) methods for structuring RWD is thus essential for scaling real-world evidence generation. Materials and Methods: Traditional rule-based systems are vulnerable to the prevalent linguistic variations and ambiguities in clinical text, and prior applications of machine-learning methods typically require sentence-level or report-level labeled examples that are hard to produce at scale. We propose leveraging patient-level supervision from medical registries, which are often readily available and capture key patient information, for general RWD applications. To combat the lack of sentence-level or report-level annotations, we explore advanced deep-learning methods by combining domain-specific pretraining, recurrent neural networks, and hierarchical attention. Results: We conduct an extensive study on 135,107 patients from the cancer registry of a large integrated delivery network (IDN) comprising healthcare systems in five western US states. Our deep learning methods attain test AUROC of 94-99% for key tumor attributes and comparable performance on held-out data from separate health systems and states. Discussion and Conclusion: Ablation results demonstrate clear superiority of these advanced deep-learning methods over prior approaches. Error analysis shows that our NLP system sometimes even corrects errors in registrar labels. We also conduct a preliminary investigation in accelerating registry curation and general RWD structuring via assisted curation for over 1.2 million cancer patients in this healthcare network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge