Theodore Zhao
Boltzmann Attention Sampling for Image Analysis with Small Objects
Mar 04, 2025Abstract:Detecting and segmenting small objects, such as lung nodules and tumor lesions, remains a critical challenge in image analysis. These objects often occupy less than 0.1% of an image, making traditional transformer architectures inefficient and prone to performance degradation due to redundant attention computations on irrelevant regions. Existing sparse attention mechanisms rely on rigid hierarchical structures, which are poorly suited for detecting small, variable, and uncertain object locations. In this paper, we propose BoltzFormer, a novel transformer-based architecture designed to address these challenges through dynamic sparse attention. BoltzFormer identifies and focuses attention on relevant areas by modeling uncertainty using a Boltzmann distribution with an annealing schedule. Initially, a higher temperature allows broader area sampling in early layers, when object location uncertainty is greatest. As the temperature decreases in later layers, attention becomes more focused, enhancing efficiency and accuracy. BoltzFormer seamlessly integrates into existing transformer architectures via a modular Boltzmann attention sampling mechanism. Comprehensive evaluations on benchmark datasets demonstrate that BoltzFormer significantly improves segmentation performance for small objects while reducing attention computation by an order of magnitude compared to previous state-of-the-art methods.
Universal Abstraction: Harnessing Frontier Models to Structure Real-World Data at Scale
Feb 02, 2025



Abstract:The vast majority of real-world patient information resides in unstructured clinical text, and the process of medical abstraction seeks to extract and normalize structured information from this unstructured input. However, traditional medical abstraction methods can require significant manual efforts that can include crafting rules or annotating training labels, limiting scalability. In this paper, we propose UniMedAbstractor (UMA), a zero-shot medical abstraction framework leveraging Large Language Models (LLMs) through a modular and customizable prompt template. We refer to our approach as universal abstraction as it can quickly scale to new attributes through its universal prompt template without curating attribute-specific training labels or rules. We evaluate UMA for oncology applications, focusing on fifteen key attributes representing the cancer patient journey, from short-context attributes (e.g., performance status, treatment) to complex long-context attributes requiring longitudinal reasoning (e.g., tumor site, histology, TNM staging). Experiments on real-world data show UMA's strong performance and generalizability. Compared to supervised and heuristic baselines, UMA with GPT-4o achieves on average an absolute 2-point F1/accuracy improvement for both short-context and long-context attribute abstraction. For pathologic T staging, UMA even outperforms the supervised model by 20 points in accuracy.
BiomedParse: a biomedical foundation model for image parsing of everything everywhere all at once
May 21, 2024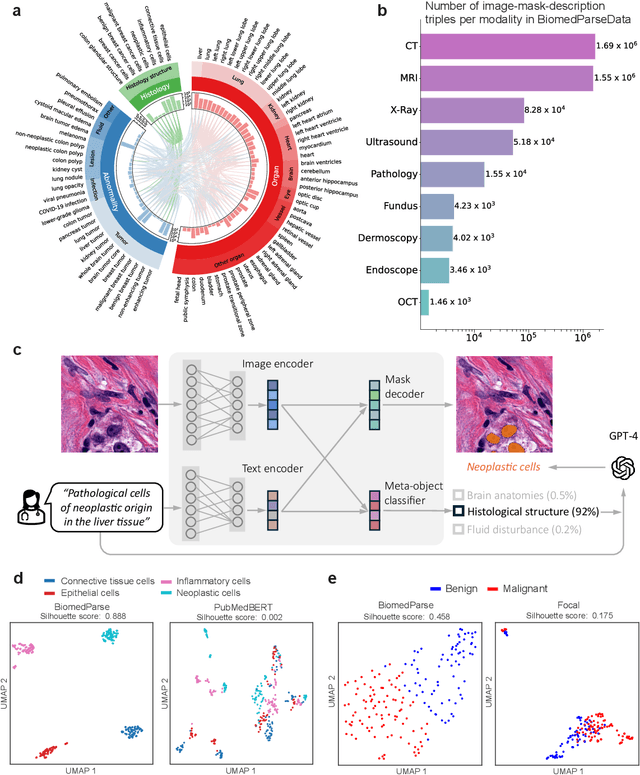
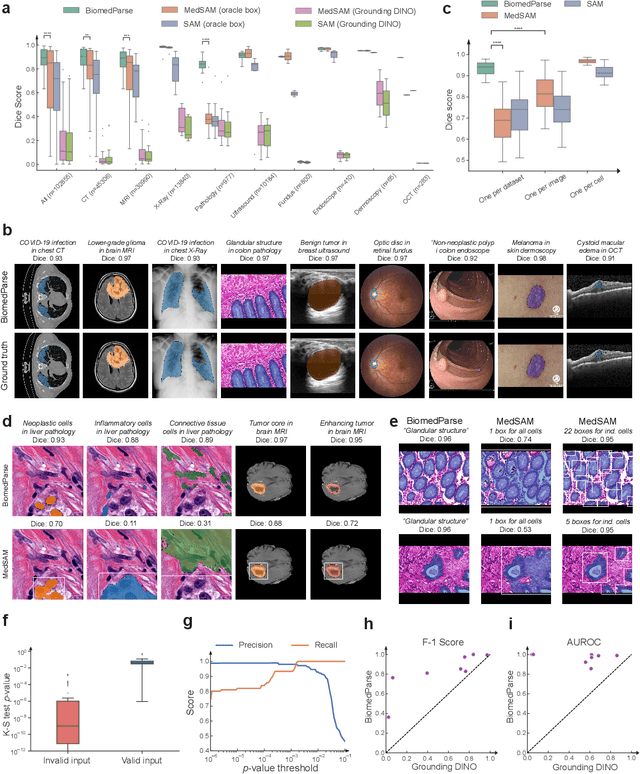
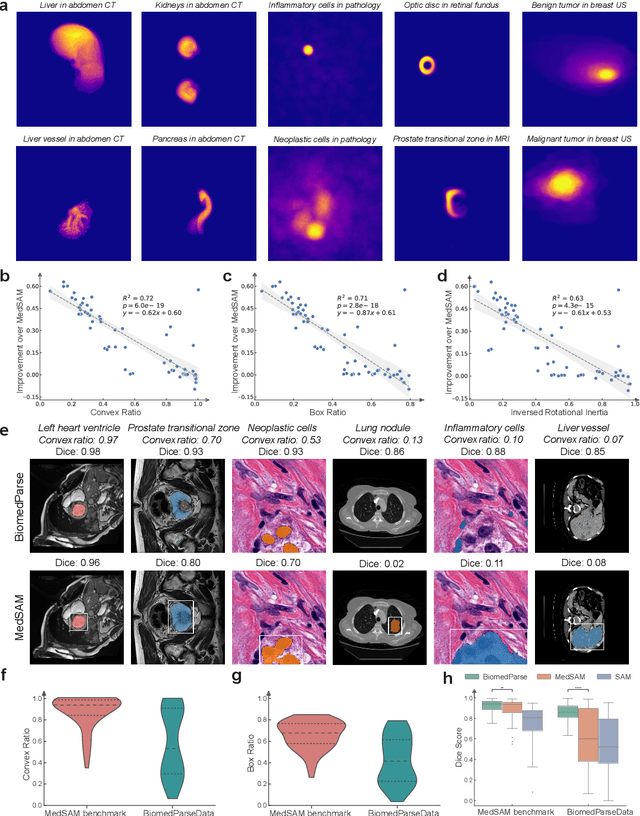
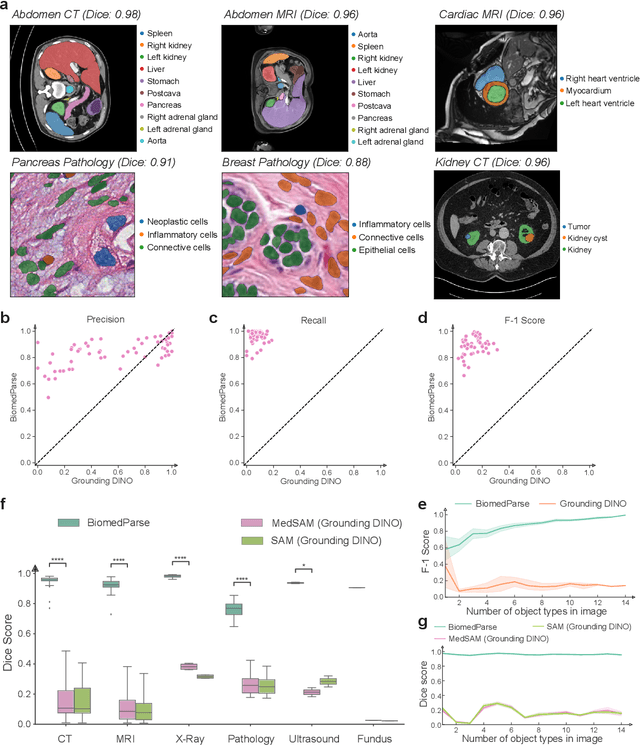
Abstract:Biomedical image analysis is fundamental for biomedical discovery in cell biology, pathology, radiology, and many other biomedical domains. Holistic image analysis comprises interdependent subtasks such as segmentation, detection, and recognition of relevant objects. Here, we propose BiomedParse, a biomedical foundation model for imaging parsing that can jointly conduct segmentation, detection, and recognition for 82 object types across 9 imaging modalities. Through joint learning, we can improve accuracy for individual tasks and enable novel applications such as segmenting all relevant objects in an image through a text prompt, rather than requiring users to laboriously specify the bounding box for each object. We leveraged readily available natural-language labels or descriptions accompanying those datasets and use GPT-4 to harmonize the noisy, unstructured text information with established biomedical object ontologies. We created a large dataset comprising over six million triples of image, segmentation mask, and textual description. On image segmentation, we showed that BiomedParse is broadly applicable, outperforming state-of-the-art methods on 102,855 test image-mask-label triples across 9 imaging modalities (everything). On object detection, which aims to locate a specific object of interest, BiomedParse again attained state-of-the-art performance, especially on objects with irregular shapes (everywhere). On object recognition, which aims to identify all objects in a given image along with their semantic types, we showed that BiomedParse can simultaneously segment and label all biomedical objects in an image (all at once). In summary, BiomedParse is an all-in-one tool for biomedical image analysis by jointly solving segmentation, detection, and recognition for all major biomedical image modalities, paving the path for efficient and accurate image-based biomedical discovery.
Foundation Models for Biomedical Image Segmentation: A Survey
Jan 15, 2024Abstract:Recent advancements in biomedical image analysis have been significantly driven by the Segment Anything Model (SAM). This transformative technology, originally developed for general-purpose computer vision, has found rapid application in medical image processing. Within the last year, marked by over 100 publications, SAM has demonstrated its prowess in zero-shot learning adaptations for medical imaging. The fundamental premise of SAM lies in its capability to segment or identify objects in images without prior knowledge of the object type or imaging modality. This approach aligns well with tasks achievable by the human visual system, though its application in non-biological vision contexts remains more theoretically challenging. A notable feature of SAM is its ability to adjust segmentation according to a specified resolution scale or area of interest, akin to semantic priming. This adaptability has spurred a wave of creativity and innovation in applying SAM to medical imaging. Our review focuses on the period from April 1, 2023, to September 30, 2023, a critical first six months post-initial publication. We examine the adaptations and integrations of SAM necessary to address longstanding clinical challenges, particularly in the context of 33 open datasets covered in our analysis. While SAM approaches or achieves state-of-the-art performance in numerous applications, it falls short in certain areas, such as segmentation of the carotid artery, adrenal glands, optic nerve, and mandible bone. Our survey delves into the innovative techniques where SAM's foundational approach excels and explores the core concepts in translating and applying these models effectively in diverse medical imaging scenarios.
LLM Calibration and Automatic Hallucination Detection via Pareto Optimal Self-supervision
Jul 06, 2023



Abstract:Large language models (LLMs) have demonstrated remarkable capabilities out of box for a wide range of applications, yet accuracy still remains a major growth area, especially in mission-critical domains such as biomedicine. An effective method to calibrate the confidence level on LLM responses is essential to automatically detect errors and facilitate human-in-the-loop verification. An important source of calibration signals stems from expert-stipulated programmatic supervision, which is often available at low cost but has its own limitations such as noise and coverage. In this paper, we introduce a Pareto optimal self-supervision framework that can leverage available programmatic supervision to systematically calibrate LLM responses by producing a risk score for every response, without any additional manual efforts. This is accomplished by learning a harmonizer model to align LLM output with other available supervision sources, which would assign higher risk scores to more uncertain LLM responses and facilitate error correction. Experiments on standard relation extraction tasks in biomedical and general domains demonstrate the promise of this approach, with our proposed risk scores highly correlated with the real error rate of LLMs. For the most uncertain test instances, dynamic prompting based on our proposed risk scores results in significant accuracy improvement for off-the-shelf LLMs, boosting GPT-3 results past state-of-the-art (SOTA) weak supervision and GPT-4 results past SOTA supervised results on challenging evaluation datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge