Hoifung Poon
Learning Sparse Visual Representations via Spatial-Semantic Factorization
Feb 02, 2026Abstract:Self-supervised learning (SSL) faces a fundamental conflict between semantic understanding and image reconstruction. High-level semantic SSL (e.g., DINO) relies on global tokens that are forced to be location-invariant for augmentation alignment, a process that inherently discards the spatial coordinates required for reconstruction. Conversely, generative SSL (e.g., MAE) preserves dense feature grids for reconstruction but fails to produce high-level abstractions. We introduce STELLAR, a framework that resolves this tension by factorizing visual features into a low-rank product of semantic concepts and their spatial distributions. This disentanglement allows us to perform DINO-style augmentation alignment on the semantic tokens while maintaining the precise spatial mapping in the localization matrix necessary for pixel-level reconstruction. We demonstrate that as few as 16 sparse tokens under this factorized form are sufficient to simultaneously support high-quality reconstruction (2.60 FID) and match the semantic performance of dense backbones (79.10% ImageNet accuracy). Our results highlight STELLAR as a versatile sparse representation that bridges the gap between discriminative and generative vision by strategically separating semantic identity from spatial geometry. Code available at https://aka.ms/stellar.
Scaling medical imaging report generation with multimodal reinforcement learning
Jan 23, 2026Abstract:Frontier models have demonstrated remarkable capabilities in understanding and reasoning with natural-language text, but they still exhibit major competency gaps in multimodal understanding and reasoning especially in high-value verticals such as biomedicine. Medical imaging report generation is a prominent example. Supervised fine-tuning can substantially improve performance, but they are prone to overfitting to superficial boilerplate patterns. In this paper, we introduce Universal Report Generation (UniRG) as a general framework for medical imaging report generation. By leveraging reinforcement learning as a unifying mechanism to directly optimize for evaluation metrics designed for end applications, UniRG can significantly improve upon supervised fine-tuning and attain durable generalization across diverse institutions and clinical practices. We trained UniRG-CXR on publicly available chest X-ray (CXR) data and conducted a thorough evaluation in CXR report generation with rigorous evaluation scenarios. On the authoritative ReXrank benchmark, UniRG-CXR sets new overall SOTA, outperforming prior state of the art by a wide margin.
ArenaBencher: Automatic Benchmark Evolution via Multi-Model Competitive Evaluation
Oct 09, 2025
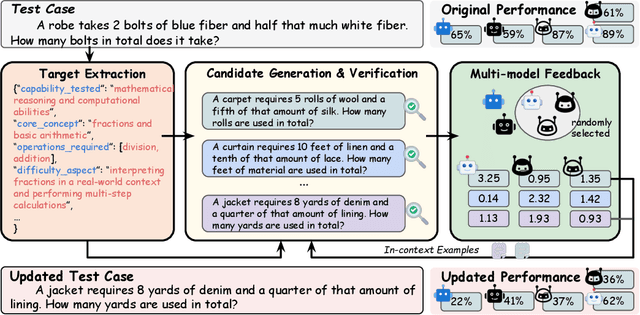
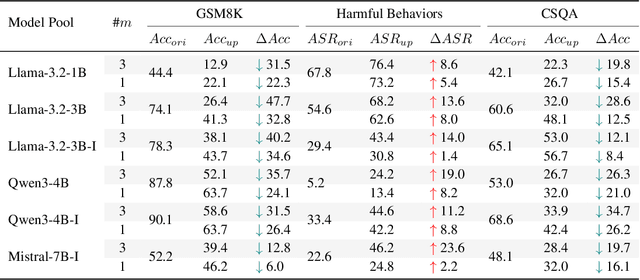
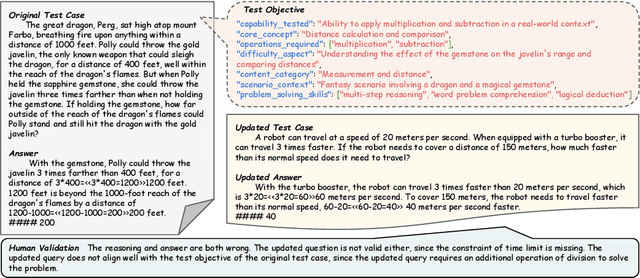
Abstract:Benchmarks are central to measuring the capabilities of large language models and guiding model development, yet widespread data leakage from pretraining corpora undermines their validity. Models can match memorized content rather than demonstrate true generalization, which inflates scores, distorts cross-model comparisons, and misrepresents progress. We introduce ArenaBencher, a model-agnostic framework for automatic benchmark evolution that updates test cases while preserving comparability. Given an existing benchmark and a diverse pool of models to be evaluated, ArenaBencher infers the core ability of each test case, generates candidate question-answer pairs that preserve the original objective, verifies correctness and intent with an LLM as a judge, and aggregates feedback from multiple models to select candidates that expose shared weaknesses. The process runs iteratively with in-context demonstrations that steer generation toward more challenging and diagnostic cases. We apply ArenaBencher to math problem solving, commonsense reasoning, and safety domains and show that it produces verified, diverse, and fair updates that uncover new failure modes, increase difficulty while preserving test objective alignment, and improve model separability. The framework provides a scalable path to continuously evolve benchmarks in step with the rapid progress of foundation models.
CancerGUIDE: Cancer Guideline Understanding via Internal Disagreement Estimation
Sep 09, 2025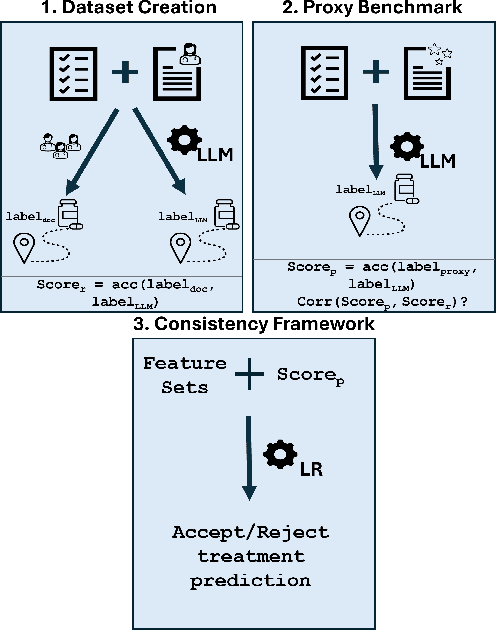
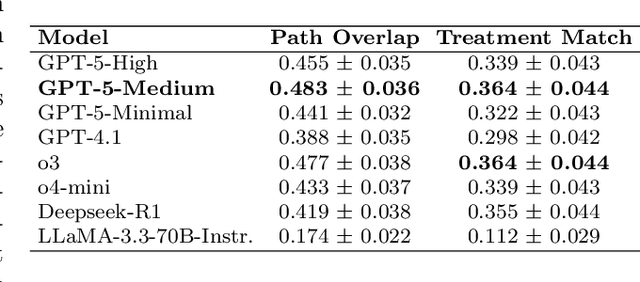
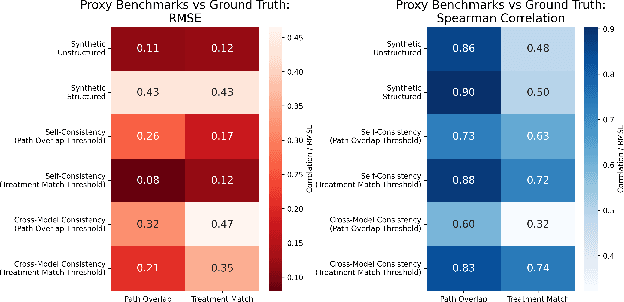
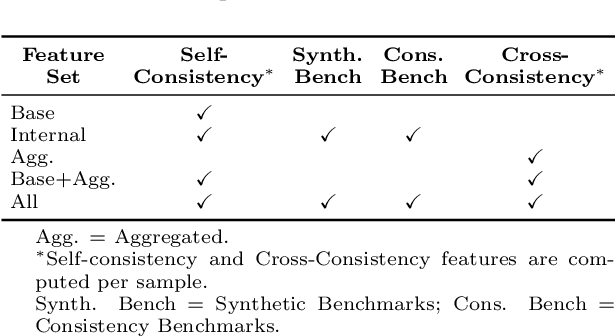
Abstract:The National Comprehensive Cancer Network (NCCN) provides evidence-based guidelines for cancer treatment. Translating complex patient presentations into guideline-compliant treatment recommendations is time-intensive, requires specialized expertise, and is prone to error. Advances in large language model (LLM) capabilities promise to reduce the time required to generate treatment recommendations and improve accuracy. We present an LLM agent-based approach to automatically generate guideline-concordant treatment trajectories for patients with non-small cell lung cancer (NSCLC). Our contributions are threefold. First, we construct a novel longitudinal dataset of 121 cases of NSCLC patients that includes clinical encounters, diagnostic results, and medical histories, each expertly annotated with the corresponding NCCN guideline trajectories by board-certified oncologists. Second, we demonstrate that existing LLMs possess domain-specific knowledge that enables high-quality proxy benchmark generation for both model development and evaluation, achieving strong correlation (Spearman coefficient r=0.88, RMSE = 0.08) with expert-annotated benchmarks. Third, we develop a hybrid approach combining expensive human annotations with model consistency information to create both the agent framework that predicts the relevant guidelines for a patient, as well as a meta-classifier that verifies prediction accuracy with calibrated confidence scores for treatment recommendations (AUROC=0.800), a critical capability for communicating the accuracy of outputs, custom-tailoring tradeoffs in performance, and supporting regulatory compliance. This work establishes a framework for clinically viable LLM-based guideline adherence systems that balance accuracy, interpretability, and regulatory requirements while reducing annotation costs, providing a scalable pathway toward automated clinical decision support.
AURAD: Anatomy-Pathology Unified Radiology Synthesis with Progressive Representations
Sep 05, 2025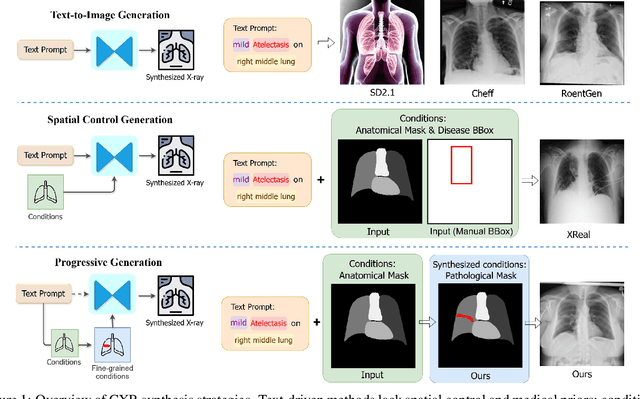

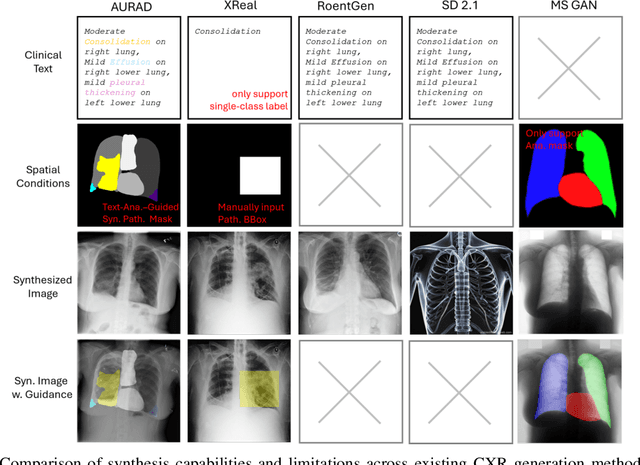
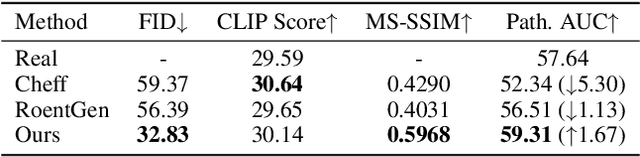
Abstract:Medical image synthesis has become an essential strategy for augmenting datasets and improving model generalization in data-scarce clinical settings. However, fine-grained and controllable synthesis remains difficult due to limited high-quality annotations and domain shifts across datasets. Existing methods, often designed for natural images or well-defined tumors, struggle to generalize to chest radiographs, where disease patterns are morphologically diverse and tightly intertwined with anatomical structures. To address these challenges, we propose AURAD, a controllable radiology synthesis framework that jointly generates high-fidelity chest X-rays and pseudo semantic masks. Unlike prior approaches that rely on randomly sampled masks-limiting diversity, controllability, and clinical relevance-our method learns to generate masks that capture multi-pathology coexistence and anatomical-pathological consistency. It follows a progressive pipeline: pseudo masks are first generated from clinical prompts conditioned on anatomical structures, and then used to guide image synthesis. We also leverage pretrained expert medical models to filter outputs and ensure clinical plausibility. Beyond visual realism, the synthesized masks also serve as labels for downstream tasks such as detection and segmentation, bridging the gap between generative modeling and real-world clinical applications. Extensive experiments and blinded radiologist evaluations demonstrate the effectiveness and generalizability of our method across tasks and datasets. In particular, 78% of our synthesized images are classified as authentic by board-certified radiologists, and over 40% of predicted segmentation overlays are rated as clinically useful. All code, pre-trained models, and the synthesized dataset will be released upon publication.
Exploring Scaling Laws for EHR Foundation Models
May 29, 2025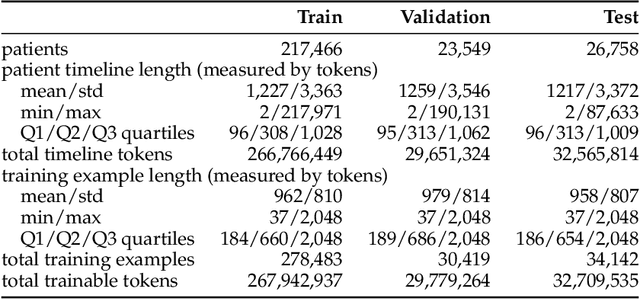
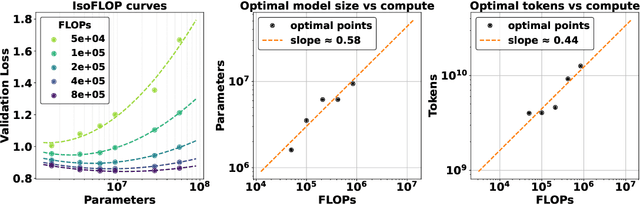
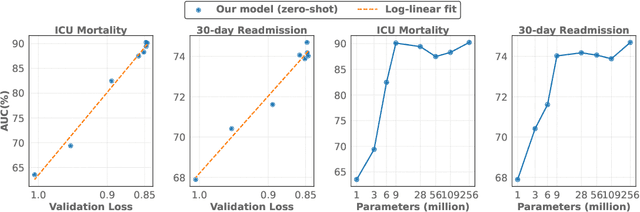
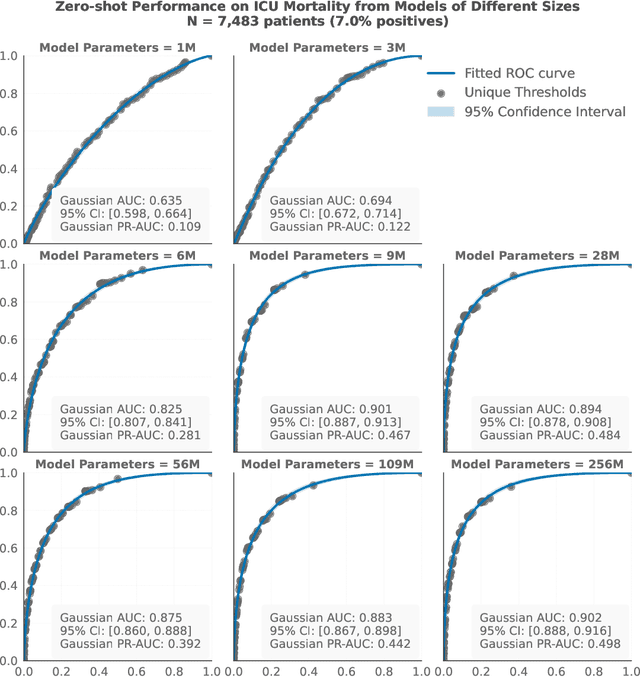
Abstract:The emergence of scaling laws has profoundly shaped the development of large language models (LLMs), enabling predictable performance gains through systematic increases in model size, dataset volume, and compute. Yet, these principles remain largely unexplored in the context of electronic health records (EHRs) -- a rich, sequential, and globally abundant data source that differs structurally from natural language. In this work, we present the first empirical investigation of scaling laws for EHR foundation models. By training transformer architectures on patient timeline data from the MIMIC-IV database across varying model sizes and compute budgets, we identify consistent scaling patterns, including parabolic IsoFLOPs curves and power-law relationships between compute, model parameters, data size, and clinical utility. These findings demonstrate that EHR models exhibit scaling behavior analogous to LLMs, offering predictive insights into resource-efficient training strategies. Our results lay the groundwork for developing powerful EHR foundation models capable of transforming clinical prediction tasks and advancing personalized healthcare.
X-Reasoner: Towards Generalizable Reasoning Across Modalities and Domains
May 06, 2025Abstract:Recent proprietary models (e.g., o3) have begun to demonstrate strong multimodal reasoning capabilities. Yet, most existing open-source research concentrates on training text-only reasoning models, with evaluations limited to mainly mathematical and general-domain tasks. Therefore, it remains unclear how to effectively extend reasoning capabilities beyond text input and general domains. This paper explores a fundamental research question: Is reasoning generalizable across modalities and domains? Our findings support an affirmative answer: General-domain text-based post-training can enable such strong generalizable reasoning. Leveraging this finding, we introduce X-Reasoner, a vision-language model post-trained solely on general-domain text for generalizable reasoning, using a two-stage approach: an initial supervised fine-tuning phase with distilled long chain-of-thoughts, followed by reinforcement learning with verifiable rewards. Experiments show that X-Reasoner successfully transfers reasoning capabilities to both multimodal and out-of-domain settings, outperforming existing state-of-the-art models trained with in-domain and multimodal data across various general and medical benchmarks (Figure 1). Additionally, we find that X-Reasoner's performance in specialized domains can be further enhanced through continued training on domain-specific text-only data. Building upon this, we introduce X-Reasoner-Med, a medical-specialized variant that achieves new state of the art on numerous text-only and multimodal medical benchmarks.
MetaScale: Test-Time Scaling with Evolving Meta-Thoughts
Mar 17, 2025Abstract:One critical challenge for large language models (LLMs) for making complex reasoning is their reliance on matching reasoning patterns from training data, instead of proactively selecting the most appropriate cognitive strategy to solve a given task. Existing approaches impose fixed cognitive structures that enhance performance in specific tasks but lack adaptability across diverse scenarios. To address this limitation, we introduce METASCALE, a test-time scaling framework based on meta-thoughts -- adaptive thinking strategies tailored to each task. METASCALE initializes a pool of candidate meta-thoughts, then iteratively selects and evaluates them using a multi-armed bandit algorithm with upper confidence bound selection, guided by a reward model. To further enhance adaptability, a genetic algorithm evolves high-reward meta-thoughts, refining and extending the strategy pool over time. By dynamically proposing and optimizing meta-thoughts at inference time, METASCALE improves both accuracy and generalization across a wide range of tasks. Experimental results demonstrate that MetaScale consistently outperforms standard inference approaches, achieving an 11% performance gain in win rate on Arena-Hard for GPT-4o, surpassing o1-mini by 0.9% under style control. Notably, METASCALE scales more effectively with increasing sampling budgets and produces more structured, expert-level responses.
Semantic-Clipping: Efficient Vision-Language Modeling with Semantic-Guidedd Visual Selection
Mar 14, 2025Abstract:Vision-Language Models (VLMs) leverage aligned visual encoders to transform images into visual tokens, allowing them to be processed similarly to text by the backbone large language model (LLM). This unified input paradigm enables VLMs to excel in vision-language tasks such as visual question answering (VQA). To improve fine-grained visual reasoning, recent advancements in vision-language modeling introduce image cropping techniques that feed all encoded sub-images into the model. However, this approach significantly increases the number of visual tokens, leading to inefficiency and potential distractions for the LLM. To address the generalization challenges of image representation in VLMs, we propose a lightweight, universal framework that seamlessly integrates with existing VLMs to enhance their ability to process finegrained details. Our method leverages textual semantics to identify key visual areas, improving VQA performance without requiring any retraining of the VLM. Additionally, it incorporates textual signals into the visual encoding process, enhancing both efficiency and effectiveness. The proposed method, SEMCLIP, strengthens the visual understanding of a 7B VLM, LLaVA-1.5 by 3.3% on average across 7 benchmarks, and particularly by 5.3% on the challenging detailed understanding benchmark V*.
Boltzmann Attention Sampling for Image Analysis with Small Objects
Mar 04, 2025Abstract:Detecting and segmenting small objects, such as lung nodules and tumor lesions, remains a critical challenge in image analysis. These objects often occupy less than 0.1% of an image, making traditional transformer architectures inefficient and prone to performance degradation due to redundant attention computations on irrelevant regions. Existing sparse attention mechanisms rely on rigid hierarchical structures, which are poorly suited for detecting small, variable, and uncertain object locations. In this paper, we propose BoltzFormer, a novel transformer-based architecture designed to address these challenges through dynamic sparse attention. BoltzFormer identifies and focuses attention on relevant areas by modeling uncertainty using a Boltzmann distribution with an annealing schedule. Initially, a higher temperature allows broader area sampling in early layers, when object location uncertainty is greatest. As the temperature decreases in later layers, attention becomes more focused, enhancing efficiency and accuracy. BoltzFormer seamlessly integrates into existing transformer architectures via a modular Boltzmann attention sampling mechanism. Comprehensive evaluations on benchmark datasets demonstrate that BoltzFormer significantly improves segmentation performance for small objects while reducing attention computation by an order of magnitude compared to previous state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge