Sid Kiblawi
Learning Sparse Visual Representations via Spatial-Semantic Factorization
Feb 02, 2026Abstract:Self-supervised learning (SSL) faces a fundamental conflict between semantic understanding and image reconstruction. High-level semantic SSL (e.g., DINO) relies on global tokens that are forced to be location-invariant for augmentation alignment, a process that inherently discards the spatial coordinates required for reconstruction. Conversely, generative SSL (e.g., MAE) preserves dense feature grids for reconstruction but fails to produce high-level abstractions. We introduce STELLAR, a framework that resolves this tension by factorizing visual features into a low-rank product of semantic concepts and their spatial distributions. This disentanglement allows us to perform DINO-style augmentation alignment on the semantic tokens while maintaining the precise spatial mapping in the localization matrix necessary for pixel-level reconstruction. We demonstrate that as few as 16 sparse tokens under this factorized form are sufficient to simultaneously support high-quality reconstruction (2.60 FID) and match the semantic performance of dense backbones (79.10% ImageNet accuracy). Our results highlight STELLAR as a versatile sparse representation that bridges the gap between discriminative and generative vision by strategically separating semantic identity from spatial geometry. Code available at https://aka.ms/stellar.
Scaling medical imaging report generation with multimodal reinforcement learning
Jan 23, 2026Abstract:Frontier models have demonstrated remarkable capabilities in understanding and reasoning with natural-language text, but they still exhibit major competency gaps in multimodal understanding and reasoning especially in high-value verticals such as biomedicine. Medical imaging report generation is a prominent example. Supervised fine-tuning can substantially improve performance, but they are prone to overfitting to superficial boilerplate patterns. In this paper, we introduce Universal Report Generation (UniRG) as a general framework for medical imaging report generation. By leveraging reinforcement learning as a unifying mechanism to directly optimize for evaluation metrics designed for end applications, UniRG can significantly improve upon supervised fine-tuning and attain durable generalization across diverse institutions and clinical practices. We trained UniRG-CXR on publicly available chest X-ray (CXR) data and conducted a thorough evaluation in CXR report generation with rigorous evaluation scenarios. On the authoritative ReXrank benchmark, UniRG-CXR sets new overall SOTA, outperforming prior state of the art by a wide margin.
X-Reasoner: Towards Generalizable Reasoning Across Modalities and Domains
May 06, 2025Abstract:Recent proprietary models (e.g., o3) have begun to demonstrate strong multimodal reasoning capabilities. Yet, most existing open-source research concentrates on training text-only reasoning models, with evaluations limited to mainly mathematical and general-domain tasks. Therefore, it remains unclear how to effectively extend reasoning capabilities beyond text input and general domains. This paper explores a fundamental research question: Is reasoning generalizable across modalities and domains? Our findings support an affirmative answer: General-domain text-based post-training can enable such strong generalizable reasoning. Leveraging this finding, we introduce X-Reasoner, a vision-language model post-trained solely on general-domain text for generalizable reasoning, using a two-stage approach: an initial supervised fine-tuning phase with distilled long chain-of-thoughts, followed by reinforcement learning with verifiable rewards. Experiments show that X-Reasoner successfully transfers reasoning capabilities to both multimodal and out-of-domain settings, outperforming existing state-of-the-art models trained with in-domain and multimodal data across various general and medical benchmarks (Figure 1). Additionally, we find that X-Reasoner's performance in specialized domains can be further enhanced through continued training on domain-specific text-only data. Building upon this, we introduce X-Reasoner-Med, a medical-specialized variant that achieves new state of the art on numerous text-only and multimodal medical benchmarks.
Boltzmann Attention Sampling for Image Analysis with Small Objects
Mar 04, 2025Abstract:Detecting and segmenting small objects, such as lung nodules and tumor lesions, remains a critical challenge in image analysis. These objects often occupy less than 0.1% of an image, making traditional transformer architectures inefficient and prone to performance degradation due to redundant attention computations on irrelevant regions. Existing sparse attention mechanisms rely on rigid hierarchical structures, which are poorly suited for detecting small, variable, and uncertain object locations. In this paper, we propose BoltzFormer, a novel transformer-based architecture designed to address these challenges through dynamic sparse attention. BoltzFormer identifies and focuses attention on relevant areas by modeling uncertainty using a Boltzmann distribution with an annealing schedule. Initially, a higher temperature allows broader area sampling in early layers, when object location uncertainty is greatest. As the temperature decreases in later layers, attention becomes more focused, enhancing efficiency and accuracy. BoltzFormer seamlessly integrates into existing transformer architectures via a modular Boltzmann attention sampling mechanism. Comprehensive evaluations on benchmark datasets demonstrate that BoltzFormer significantly improves segmentation performance for small objects while reducing attention computation by an order of magnitude compared to previous state-of-the-art methods.
Universal Abstraction: Harnessing Frontier Models to Structure Real-World Data at Scale
Feb 02, 2025



Abstract:The vast majority of real-world patient information resides in unstructured clinical text, and the process of medical abstraction seeks to extract and normalize structured information from this unstructured input. However, traditional medical abstraction methods can require significant manual efforts that can include crafting rules or annotating training labels, limiting scalability. In this paper, we propose UniMedAbstractor (UMA), a zero-shot medical abstraction framework leveraging Large Language Models (LLMs) through a modular and customizable prompt template. We refer to our approach as universal abstraction as it can quickly scale to new attributes through its universal prompt template without curating attribute-specific training labels or rules. We evaluate UMA for oncology applications, focusing on fifteen key attributes representing the cancer patient journey, from short-context attributes (e.g., performance status, treatment) to complex long-context attributes requiring longitudinal reasoning (e.g., tumor site, histology, TNM staging). Experiments on real-world data show UMA's strong performance and generalizability. Compared to supervised and heuristic baselines, UMA with GPT-4o achieves on average an absolute 2-point F1/accuracy improvement for both short-context and long-context attribute abstraction. For pathologic T staging, UMA even outperforms the supervised model by 20 points in accuracy.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022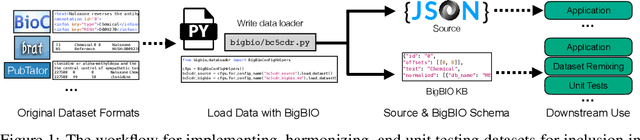
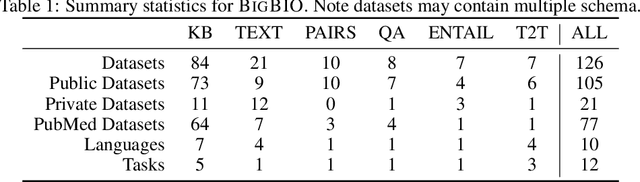
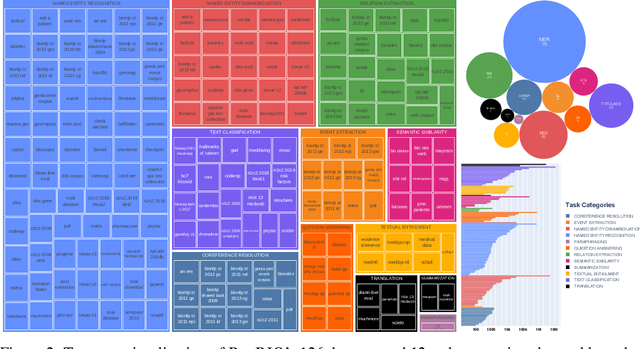
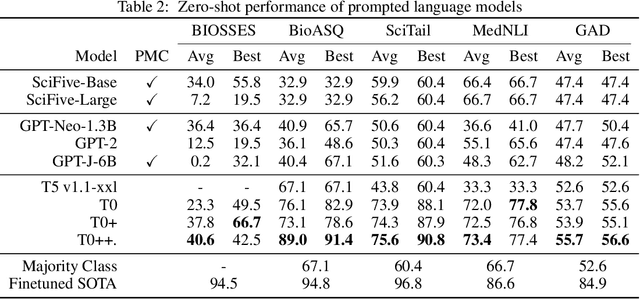
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge