Myungsun Kang
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022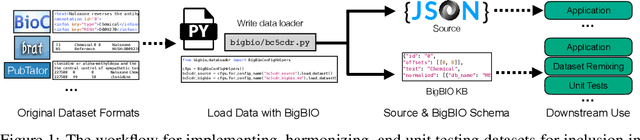
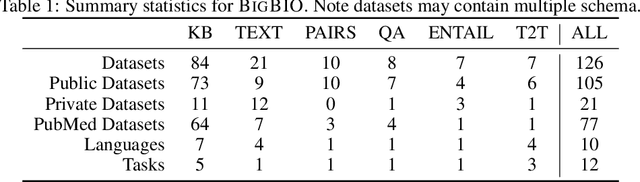
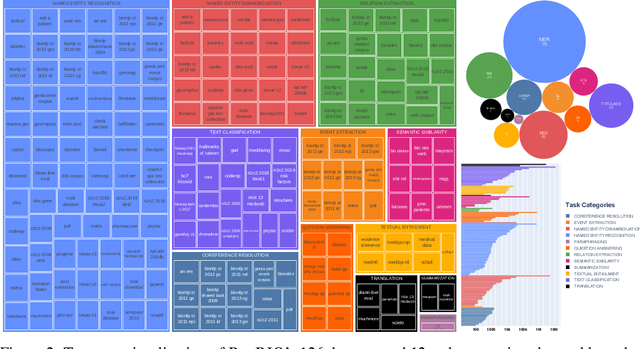
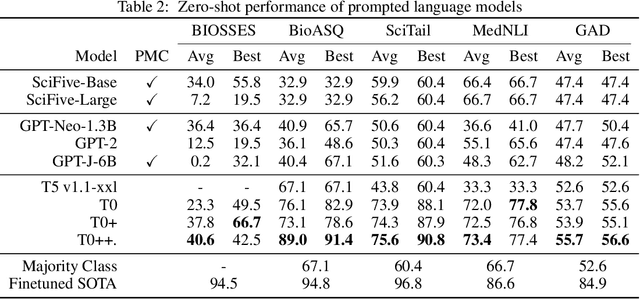
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
COVID-19 Outbreak Prediction and Analysis using Self Reported Symptoms
Dec 21, 2020
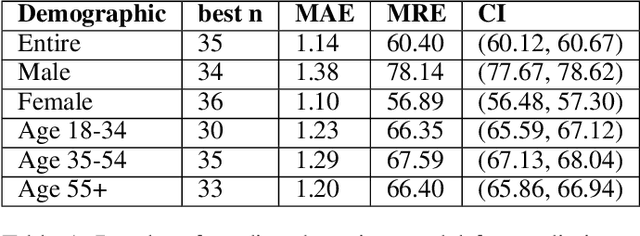
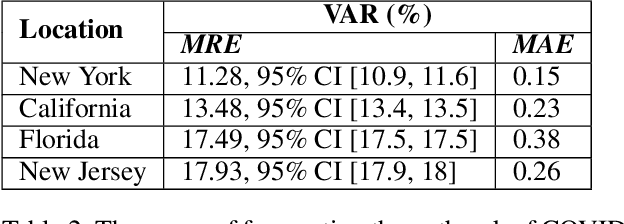

Abstract:The COVID-19 pandemic has challenged scientists and policy-makers internationally to develop novel approaches to public health policy. Furthermore, it has also been observed that the prevalence and spread of COVID-19 vary across different spatial, temporal, and demographics. Despite ramping up testing, we still are not at the required level in most parts of the globe. Therefore, we utilize self-reported symptoms survey data to understand trends in the spread of COVID-19. The aim of this study is to segment populations that are highly susceptible. In order to understand such populations, we perform exploratory data analysis, outbreak prediction, and time-series forecasting using public health and policy datasets. From our studies, we try to predict the likely % of the population that tested positive for COVID-19 based on self-reported symptoms. Our findings reaffirm the predictive value of symptoms, such as anosmia and ageusia. And we forecast that % of the population having COVID-19-like illness (CLI) and those tested positive as 0.15% and 1.14% absolute error respectively. These findings could help aid faster development of the public health policy, particularly in areas with low levels of testing and having a greater reliance on self-reported symptoms. Our analysis sheds light on identifying clinical attributes of interest across different demographics. We also provide insights into the effects of various policy enactments on COVID-19 prevalence.
Proximity Sensing for Contact Tracing
Sep 04, 2020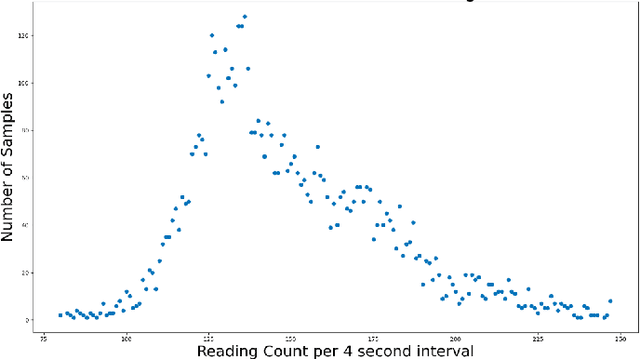

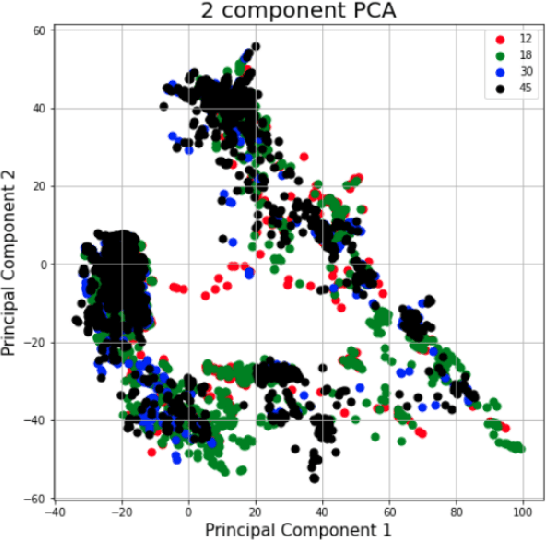

Abstract:The TC4TL (Too Close For Too Long) challenge is aimed towards designing an effective proximity sensing algorithm that can accurately provide exposure notifications. In this paper, we describe our approach to model sensor and other device-level data to estimate the distance between two phones. We also present our research and data analysis on the TC4TL challenge and discuss various limitations associated with the task, and the dataset used for this purpose.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge