Gabriel Altay
Large Language Models with Retrieval-Augmented Generation for Zero-Shot Disease Phenotyping
Dec 11, 2023Abstract:Identifying disease phenotypes from electronic health records (EHRs) is critical for numerous secondary uses. Manually encoding physician knowledge into rules is particularly challenging for rare diseases due to inadequate EHR coding, necessitating review of clinical notes. Large language models (LLMs) offer promise in text understanding but may not efficiently handle real-world clinical documentation. We propose a zero-shot LLM-based method enriched by retrieval-augmented generation and MapReduce, which pre-identifies disease-related text snippets to be used in parallel as queries for the LLM to establish diagnosis. We show that this method as applied to pulmonary hypertension (PH), a rare disease characterized by elevated arterial pressures in the lungs, significantly outperforms physician logic rules ($F_1$ score of 0.62 vs. 0.75). This method has the potential to enhance rare disease cohort identification, expanding the scope of robust clinical research and care gap identification.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022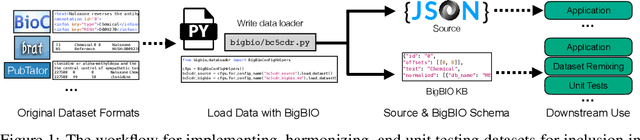
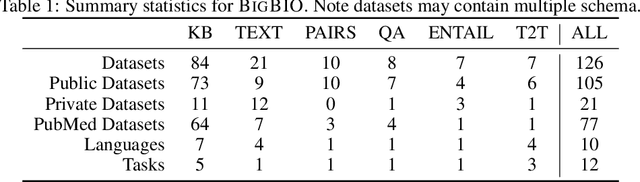
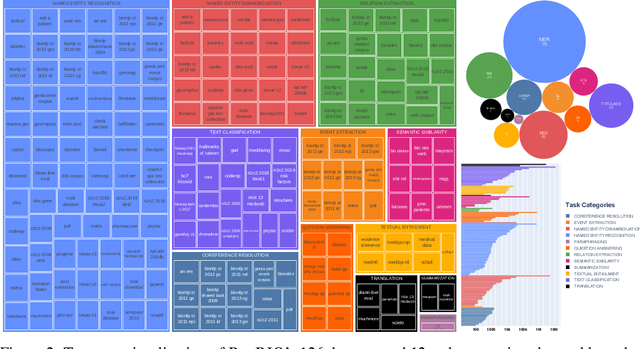
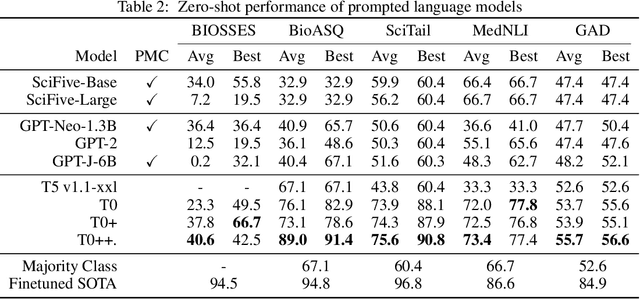
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge