Brandon K. Fornwalt
Large Language Models with Retrieval-Augmented Generation for Zero-Shot Disease Phenotyping
Dec 11, 2023Abstract:Identifying disease phenotypes from electronic health records (EHRs) is critical for numerous secondary uses. Manually encoding physician knowledge into rules is particularly challenging for rare diseases due to inadequate EHR coding, necessitating review of clinical notes. Large language models (LLMs) offer promise in text understanding but may not efficiently handle real-world clinical documentation. We propose a zero-shot LLM-based method enriched by retrieval-augmented generation and MapReduce, which pre-identifies disease-related text snippets to be used in parallel as queries for the LLM to establish diagnosis. We show that this method as applied to pulmonary hypertension (PH), a rare disease characterized by elevated arterial pressures in the lungs, significantly outperforms physician logic rules ($F_1$ score of 0.62 vs. 0.75). This method has the potential to enhance rare disease cohort identification, expanding the scope of robust clinical research and care gap identification.
Deep neural networks can predict mortality from 12-lead electrocardiogram voltage data
May 03, 2019



Abstract:The electrocardiogram (ECG) is a widely-used medical test, typically consisting of 12 voltage versus time traces collected from surface recordings over the heart. Here we hypothesize that a deep neural network can predict an important future clinical event (one-year all-cause mortality) from ECG voltage-time traces. We show good performance for predicting one-year mortality with an average AUC of 0.85 from a model cross-validated on 1,775,926 12-lead resting ECGs, that were collected over a 34-year period in a large regional health system. Even within the large subset of ECGs interpreted as 'normal' by a physician (n=297,548), the model performance to predict one-year mortality remained high (AUC=0.84), and Cox Proportional Hazard model revealed a hazard ratio of 6.6 (p<0.005) for the two predicted groups (dead vs alive one year after ECG) over a 30-year follow-up period. A blinded survey of three cardiologists suggested that the patterns captured by the model were generally not visually apparent to cardiologists even after being shown 240 paired examples of labeled true positives (dead) and true negatives (alive). In summary, deep learning can add significant prognostic information to the interpretation of 12-lead resting ECGs, even in cases that are interpreted as 'normal' by physicians.
Interpretable Neural Networks for Predicting Mortality Risk using Multi-modal Electronic Health Records
Jan 23, 2019
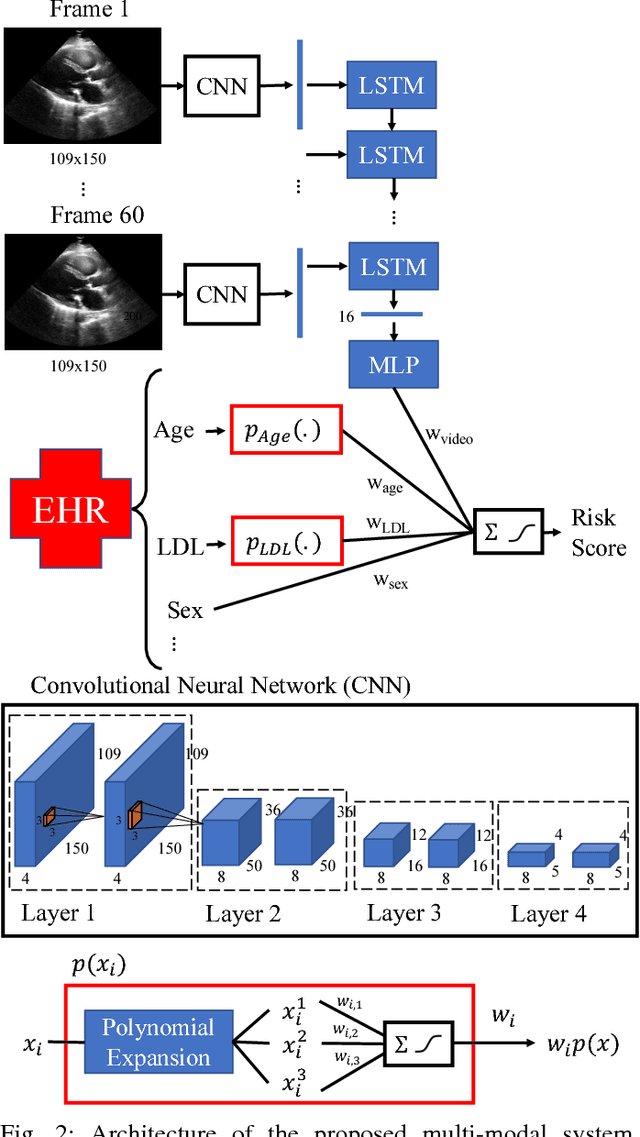

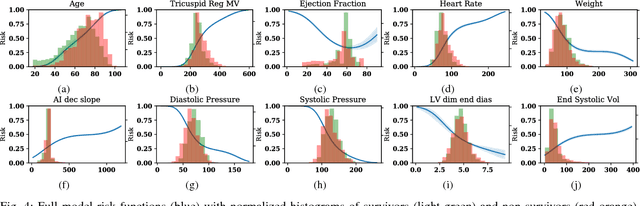
Abstract:We present an interpretable neural network for predicting an important clinical outcome (1-year mortality) from multi-modal Electronic Health Record (EHR) data. Our approach builds on prior multi-modal machine learning models by now enabling visualization of how individual factors contribute to the overall outcome risk, assuming other factors remain constant, which was previously impossible. We demonstrate the value of this approach using a large multi-modal clinical dataset including both EHR data and 31,278 echocardiographic videos of the heart from 26,793 patients. We generated separate models for (i) clinical data only (CD) (e.g. age, sex, diagnoses and laboratory values), (ii) numeric variables derived from the videos, which we call echocardiography-derived measures (EDM), and (iii) CD+EDM+raw videos (pixel data). The interpretable multi-modal model maintained performance compared to non-interpretable models (Random Forest, XGBoost), and also performed significantly better than a model using a single modality (average AUC=0.82). Clinically relevant insights and multi-modal variable importance rankings were also facilitated by the new model, which have previously been impossible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge