Christopher M. Haggerty
The Case Records of ChatGPT: Language Models and Complex Clinical Questions
May 09, 2023Abstract:Background: Artificial intelligence language models have shown promise in various applications, including assisting with clinical decision-making as demonstrated by strong performance of large language models on medical licensure exams. However, their ability to solve complex, open-ended cases, which may be representative of clinical practice, remains unexplored. Methods: In this study, the accuracy of large language AI models GPT4 and GPT3.5 in diagnosing complex clinical cases was investigated using published Case Records of the Massachusetts General Hospital. A total of 50 cases requiring a diagnosis and diagnostic test published from January 1, 2022 to April 16, 2022 were identified. For each case, models were given a prompt requesting the top three specific diagnoses and associated diagnostic tests, followed by case text, labs, and figure legends. Model outputs were assessed in comparison to the final clinical diagnosis and whether the model-predicted test would result in a correct diagnosis. Results: GPT4 and GPT3.5 accurately provided the correct diagnosis in 26% and 22% of cases in one attempt, and 46% and 42% within three attempts, respectively. GPT4 and GPT3.5 provided a correct essential diagnostic test in 28% and 24% of cases in one attempt, and 44% and 50% within three attempts, respectively. No significant differences were found between the two models, and multiple trials with identical prompts using the GPT3.5 model provided similar results. Conclusions: In summary, these models demonstrate potential usefulness in generating differential diagnoses but remain limited in their ability to provide a single unifying diagnosis in complex, open-ended cases. Future research should focus on evaluating model performance in larger datasets of open-ended clinical challenges and exploring potential human-AI collaboration strategies to enhance clinical decision-making.
Bayesian Optimization of 2D Echocardiography Segmentation
Nov 17, 2022



Abstract:Bayesian Optimization (BO) is a well-studied hyperparameter tuning technique that is more efficient than grid search for high-cost, high-parameter machine learning problems. Echocardiography is a ubiquitous modality for evaluating heart structure and function in cardiology. In this work, we use BO to optimize the architectural and training-related hyperparameters of a previously published deep fully convolutional neural network model for multi-structure segmentation in echocardiography. In a fair comparison, the resulting model outperforms this recent state-of-the-art on the annotated CAMUS dataset in both apical two- and four-chamber echo views. We report mean Dice overlaps of 0.95, 0.96, and 0.93 on left ventricular (LV) endocardium, LV epicardium, and left atrium respectively. We also observe significant improvement in derived clinical indices, including smaller median absolute errors for LV end-diastolic volume (4.9mL vs. 6.7), end-systolic volume (3.1mL vs. 5.2), and ejection fraction (2.6% vs. 3.7); and much tighter limits of agreement, which were already within inter-rater variability for non-contrast echo. These results demonstrate the benefits of BO for echocardiography segmentation over a recent state-of-the-art framework, although validation using large-scale independent clinical data is required.
Deep neural networks can predict mortality from 12-lead electrocardiogram voltage data
May 03, 2019



Abstract:The electrocardiogram (ECG) is a widely-used medical test, typically consisting of 12 voltage versus time traces collected from surface recordings over the heart. Here we hypothesize that a deep neural network can predict an important future clinical event (one-year all-cause mortality) from ECG voltage-time traces. We show good performance for predicting one-year mortality with an average AUC of 0.85 from a model cross-validated on 1,775,926 12-lead resting ECGs, that were collected over a 34-year period in a large regional health system. Even within the large subset of ECGs interpreted as 'normal' by a physician (n=297,548), the model performance to predict one-year mortality remained high (AUC=0.84), and Cox Proportional Hazard model revealed a hazard ratio of 6.6 (p<0.005) for the two predicted groups (dead vs alive one year after ECG) over a 30-year follow-up period. A blinded survey of three cardiologists suggested that the patterns captured by the model were generally not visually apparent to cardiologists even after being shown 240 paired examples of labeled true positives (dead) and true negatives (alive). In summary, deep learning can add significant prognostic information to the interpretation of 12-lead resting ECGs, even in cases that are interpreted as 'normal' by physicians.
Interpretable Neural Networks for Predicting Mortality Risk using Multi-modal Electronic Health Records
Jan 23, 2019
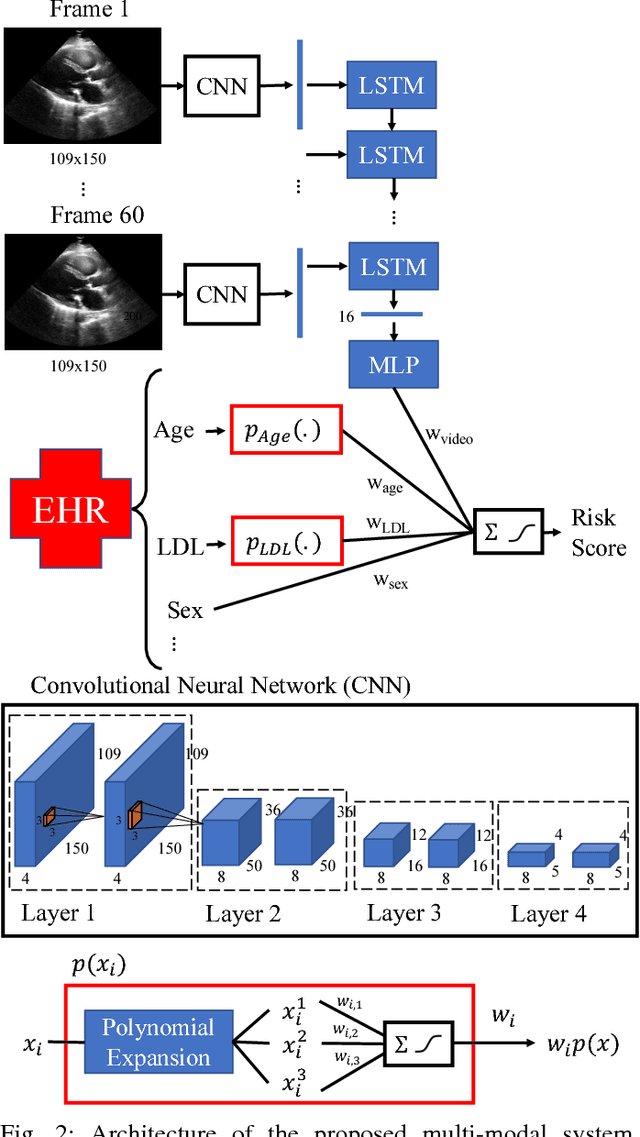

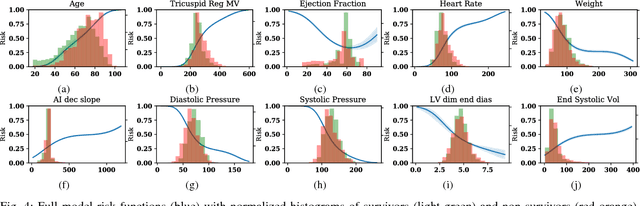
Abstract:We present an interpretable neural network for predicting an important clinical outcome (1-year mortality) from multi-modal Electronic Health Record (EHR) data. Our approach builds on prior multi-modal machine learning models by now enabling visualization of how individual factors contribute to the overall outcome risk, assuming other factors remain constant, which was previously impossible. We demonstrate the value of this approach using a large multi-modal clinical dataset including both EHR data and 31,278 echocardiographic videos of the heart from 26,793 patients. We generated separate models for (i) clinical data only (CD) (e.g. age, sex, diagnoses and laboratory values), (ii) numeric variables derived from the videos, which we call echocardiography-derived measures (EDM), and (iii) CD+EDM+raw videos (pixel data). The interpretable multi-modal model maintained performance compared to non-interpretable models (Random Forest, XGBoost), and also performed significantly better than a model using a single modality (average AUC=0.82). Clinically relevant insights and multi-modal variable importance rankings were also facilitated by the new model, which have previously been impossible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge