Joseph Bae
Token Sparsification for Faster Medical Image Segmentation
Mar 11, 2023



Abstract:Can we use sparse tokens for dense prediction, e.g., segmentation? Although token sparsification has been applied to Vision Transformers (ViT) to accelerate classification, it is still unknown how to perform segmentation from sparse tokens. To this end, we reformulate segmentation as a sparse encoding -> token completion -> dense decoding (SCD) pipeline. We first empirically show that naively applying existing approaches from classification token pruning and masked image modeling (MIM) leads to failure and inefficient training caused by inappropriate sampling algorithms and the low quality of the restored dense features. In this paper, we propose Soft-topK Token Pruning (STP) and Multi-layer Token Assembly (MTA) to address these problems. In sparse encoding, STP predicts token importance scores with a lightweight sub-network and samples the topK tokens. The intractable topK gradients are approximated through a continuous perturbed score distribution. In token completion, MTA restores a full token sequence by assembling both sparse output tokens and pruned multi-layer intermediate ones. The last dense decoding stage is compatible with existing segmentation decoders, e.g., UNETR. Experiments show SCD pipelines equipped with STP and MTA are much faster than baselines without token pruning in both training (up to 120% higher throughput and inference up to 60.6% higher throughput) while maintaining segmentation quality.
Enhancing Modality-Agnostic Representations via Meta-Learning for Brain Tumor Segmentation
Feb 08, 2023



Abstract:In the medical vision domain, different imaging modalities provide complementary information. However, in practice, not all modalities may be available during inference. Previous approaches, e.g., knowledge distillation or image synthesis, often assume the availability of full modalities for all patients during training; this is unrealistic and impractical owing to the variability in data collection across sites. We propose a novel approach to learn enhanced modality-agnostic representations by employing a novel meta-learning strategy in training, even when only a fraction of full modality patients are available. Meta-learning enhances partial modality representations to full modality representations by meta-training on partial modality data and meta-testing on limited full modality samples. Additionally, we co-supervise this feature enrichment by introducing an auxiliary adversarial learning branch. More specifically, a missing modality detector is used as a discriminator to mimic the full modality setting. Our segmentation framework significantly outperforms state-of-the-art brain tumor segmentation techniques in missing modality scenarios, as demonstrated on two brain tumor MRI datasets.
Temporal Context Matters: Enhancing Single Image Prediction with Disease Progression Representations
Mar 31, 2022



Abstract:Clinical outcome or severity prediction from medical images has largely focused on learning representations from single-timepoint or snapshot scans. It has been shown that disease progression can be better characterized by temporal imaging. We therefore hypothesized that outcome predictions can be improved by utilizing the disease progression information from sequential images. We present a deep learning approach that leverages temporal progression information to improve clinical outcome predictions from single-timepoint images. In our method, a self-attention based Temporal Convolutional Network (TCN) is used to learn a representation that is most reflective of the disease trajectory. Meanwhile, a Vision Transformer is pretrained in a self-supervised fashion to extract features from single-timepoint images. The key contribution is to design a recalibration module that employs maximum mean discrepancy loss (MMD) to align distributions of the above two contextual representations. We train our system to predict clinical outcomes and severity grades from single-timepoint images. Experiments on chest and osteoarthritis radiography datasets demonstrate that our approach outperforms other state-of-the-art techniques.
Self Pre-training with Masked Autoencoders for Medical Image Analysis
Mar 10, 2022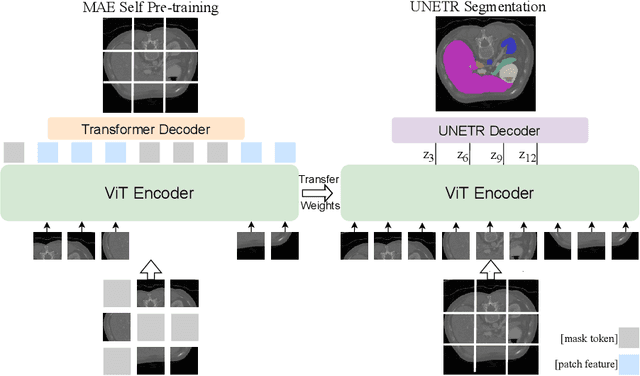
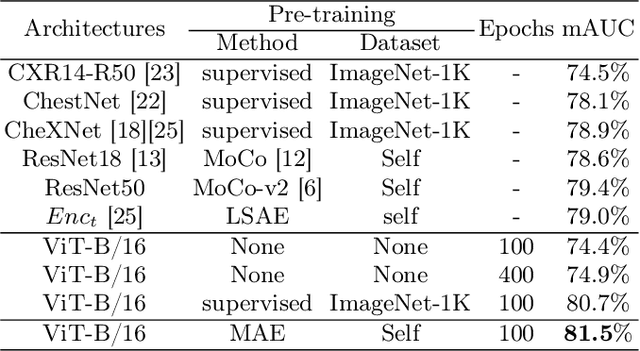
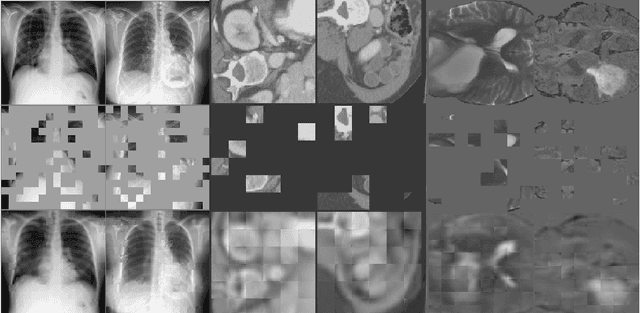
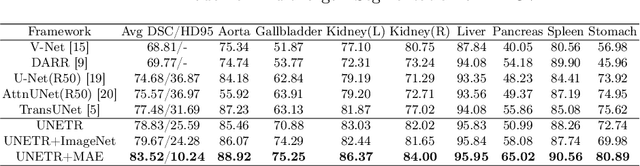
Abstract:Masked Autoencoder (MAE) has recently been shown to be effective in pre-training Vision Transformers (ViT) for natural image analysis. By performing the pretext task of reconstructing the original image from only partial observations, the encoder, which is a ViT, is encouraged to aggregate contextual information to infer content in masked image regions. We believe that this context aggregation ability is also essential to the medical image domain where each anatomical structure is functionally and mechanically connected to other structures and regions. However, there is no ImageNet-scale medical image dataset for pre-training. Thus, in this paper, we investigate a self pre-training paradigm with MAE for medical images, i.e., models are pre-trained on the same target dataset. To validate the MAE self pre-training, we consider three diverse medical image tasks including chest X-ray disease classification, CT abdomen multi-organ segmentation and MRI brain tumor segmentation. It turns out MAE self pre-training benefits all the tasks markedly. Specifically, the mAUC on lung disease classification is increased by 9.4%. The average DSC on brain tumor segmentation is improved from 77.4% to 78.9%. Most interestingly, on the small-scale multi-organ segmentation dataset (N=30), the average DSC improves from 78.8% to 83.5% and the HD95 is reduced by 60%, indicating its effectiveness in limited data scenarios. The segmentation and classification results reveal the promising potential of MAE self pre-training for medical image analysis.
Lung Swapping Autoencoder: Learning a Disentangled Structure-texture Representation of Chest Radiographs
Jan 18, 2022


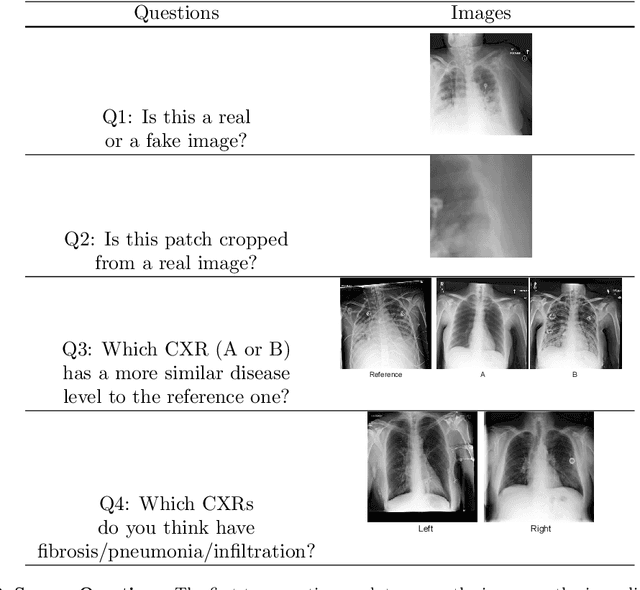
Abstract:Well-labeled datasets of chest radiographs (CXRs) are difficult to acquire due to the high cost of annotation. Thus, it is desirable to learn a robust and transferable representation in an unsupervised manner to benefit tasks that lack labeled data. Unlike natural images, medical images have their own domain prior; e.g., we observe that many pulmonary diseases, such as the COVID-19, manifest as changes in the lung tissue texture rather than the anatomical structure. Therefore, we hypothesize that studying only the texture without the influence of structure variations would be advantageous for downstream prognostic and predictive modeling tasks. In this paper, we propose a generative framework, the Lung Swapping Autoencoder (LSAE), that learns factorized representations of a CXR to disentangle the texture factor from the structure factor. Specifically, by adversarial training, the LSAE is optimized to generate a hybrid image that preserves the lung shape in one image but inherits the lung texture of another. To demonstrate the effectiveness of the disentangled texture representation, we evaluate the texture encoder $Enc^t$ in LSAE on ChestX-ray14 (N=112,120), and our own multi-institutional COVID-19 outcome prediction dataset, COVOC (N=340 (Subset-1) + 53 (Subset-2)). On both datasets, we reach or surpass the state-of-the-art by finetuning $Enc^t$ in LSAE that is 77% smaller than a baseline Inception v3. Additionally, in semi-and-self supervised settings with a similar model budget, $Enc^t$ in LSAE is also competitive with the state-of-the-art MoCo. By "re-mixing" the texture and shape factors, we generate meaningful hybrid images that can augment the training set. This data augmentation method can further improve COVOC prediction performance. The improvement is consistent even when we directly evaluate the Subset-1 trained model on Subset-2 without any fine-tuning.
Attention-based Multi-scale Gated Recurrent Encoder with Novel Correlation Loss for COVID-19 Progression Prediction
Jul 18, 2021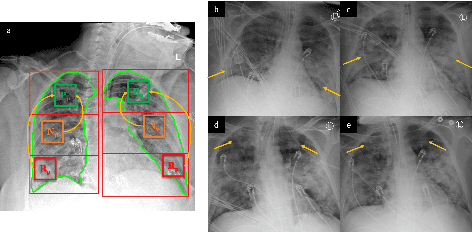


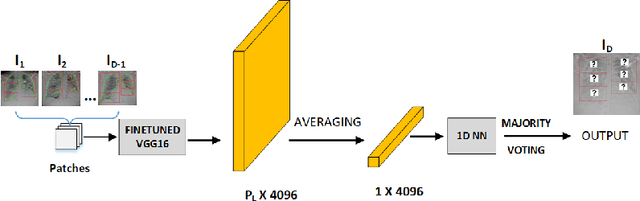
Abstract:COVID-19 image analysis has mostly focused on diagnostic tasks using single timepoint scans acquired upon disease presentation or admission. We present a deep learning-based approach to predict lung infiltrate progression from serial chest radiographs (CXRs) of COVID-19 patients. Our method first utilizes convolutional neural networks (CNNs) for feature extraction from patches within the concerned lung zone, and also from neighboring and remote boundary regions. The framework further incorporates a multi-scale Gated Recurrent Unit (GRU) with a correlation module for effective predictions. The GRU accepts CNN feature vectors from three different areas as input and generates a fused representation. The correlation module attempts to minimize the correlation loss between hidden representations of concerned and neighboring area feature vectors, while maximizing the loss between the same from concerned and remote regions. Further, we employ an attention module over the output hidden states of each encoder timepoint to generate a context vector. This vector is used as an input to a decoder module to predict patch severity grades at a future timepoint. Finally, we ensemble the patch classification scores to calculate patient-wise grades. Specifically, our framework predicts zone-wise disease severity for a patient on a given day by learning representations from the previous temporal CXRs. Our novel multi-institutional dataset comprises sequential CXR scans from N=93 patients. Our approach outperforms transfer learning and radiomic feature-based baseline approaches on this dataset.
COVID-19 Outbreak Prediction and Analysis using Self Reported Symptoms
Dec 21, 2020
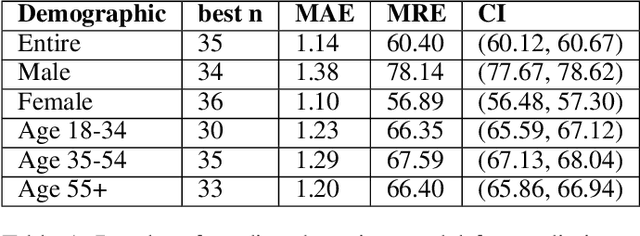
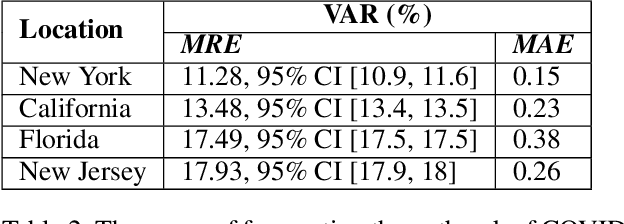

Abstract:The COVID-19 pandemic has challenged scientists and policy-makers internationally to develop novel approaches to public health policy. Furthermore, it has also been observed that the prevalence and spread of COVID-19 vary across different spatial, temporal, and demographics. Despite ramping up testing, we still are not at the required level in most parts of the globe. Therefore, we utilize self-reported symptoms survey data to understand trends in the spread of COVID-19. The aim of this study is to segment populations that are highly susceptible. In order to understand such populations, we perform exploratory data analysis, outbreak prediction, and time-series forecasting using public health and policy datasets. From our studies, we try to predict the likely % of the population that tested positive for COVID-19 based on self-reported symptoms. Our findings reaffirm the predictive value of symptoms, such as anosmia and ageusia. And we forecast that % of the population having COVID-19-like illness (CLI) and those tested positive as 0.15% and 1.14% absolute error respectively. These findings could help aid faster development of the public health policy, particularly in areas with low levels of testing and having a greater reliance on self-reported symptoms. Our analysis sheds light on identifying clinical attributes of interest across different demographics. We also provide insights into the effects of various policy enactments on COVID-19 prevalence.
Predicting Mechanical Ventilation Requirement and Mortality in COVID-19 using Radiomics and Deep Learning on Chest Radiographs: A Multi-Institutional Study
Jul 15, 2020
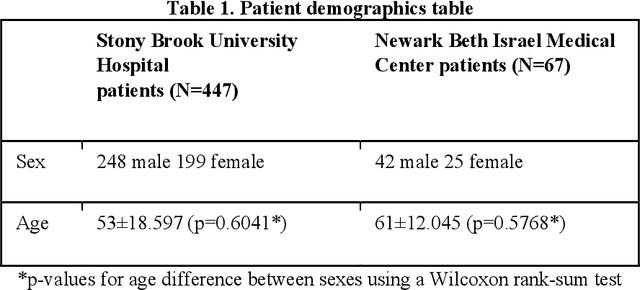

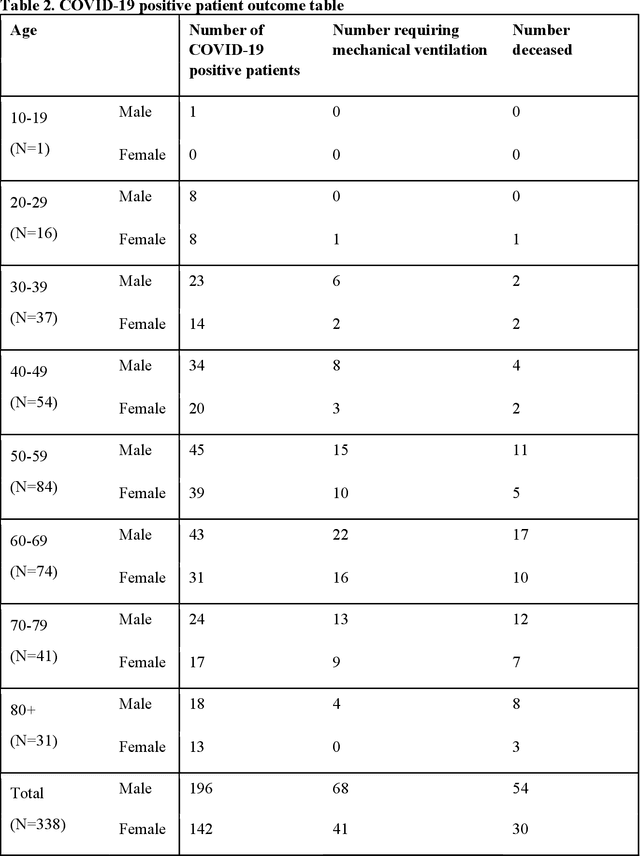
Abstract:Objectives: To predict mechanical ventilation requirement and mortality using computational modeling of chest radiographs (CXR) for coronavirus disease 2019 (COVID-19) patients. We also investigate the relative advantages of deep learning (DL), radiomics, and DL of radiomic-embedded feature maps in predicting these outcomes. Methods: This two-center, retrospective study analyzed deidentified CXRs taken from 514 patients suspected of COVID-19 infection on presentation at Stony Brook University Hospital (SBUH) and Newark Beth Israel Medical Center (NBIMC) between the months of March and June 2020. A DL segmentation pipeline was developed to generate masks for both lung fields and artifacts for each CXR. Machine learning classifiers to predict mechanical ventilation requirement and mortality were trained and evaluated on 353 baseline CXRs taken from COVID-19 positive patients. A novel radiomic embedding framework is also explored for outcome prediction. Results: Classification models for mechanical ventilation requirement (test N=154) and mortality (test N=190) had AUCs of up to 0.904 and 0.936, respectively. We also found that the inclusion of radiomic-embedded maps improved DL model predictions of clinical outcomes. Conclusions: We demonstrate the potential for computerized analysis of baseline CXR in predicting disease outcomes in COVID-19 patients. Our results also suggest that radiomic embedding improves DL models in medical image analysis, a technique that might be explored further in other pathologies. The models proposed in this study and the prognostic information they provide, complementary to other clinical data, might be used to aid physician decision making and resource allocation during the COVID-19 pandemic.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge