Amit Gupta
Multi-RADS Synthetic Radiology Report Dataset and Head-to-Head Benchmarking of 41 Open-Weight and Proprietary Language Models
Jan 06, 2026Abstract:Background: Reporting and Data Systems (RADS) standardize radiology risk communication but automated RADS assignment from narrative reports is challenging because of guideline complexity, output-format constraints, and limited benchmarking across RADS frameworks and model sizes. Purpose: To create RXL-RADSet, a radiologist-verified synthetic multi-RADS benchmark, and compare validity and accuracy of open-weight small language models (SLMs) with a proprietary model for RADS assignment. Materials and Methods: RXL-RADSet contains 1,600 synthetic radiology reports across 10 RADS (BI-RADS, CAD-RADS, GB-RADS, LI-RADS, Lung-RADS, NI-RADS, O-RADS, PI-RADS, TI-RADS, VI-RADS) and multiple modalities. Reports were generated by LLMs using scenario plans and simulated radiologist styles and underwent two-stage radiologist verification. We evaluated 41 quantized SLMs (12 families, 0.135-32B parameters) and GPT-5.2 under a fixed guided prompt. Primary endpoints were validity and accuracy; a secondary analysis compared guided versus zero-shot prompting. Results: Under guided prompting GPT-5.2 achieved 99.8% validity and 81.1% accuracy (1,600 predictions). Pooled SLMs (65,600 predictions) achieved 96.8% validity and 61.1% accuracy; top SLMs in the 20-32B range reached ~99% validity and mid-to-high 70% accuracy. Performance scaled with model size (inflection between <1B and >=10B) and declined with RADS complexity primarily due to classification difficulty rather than invalid outputs. Guided prompting improved validity (99.2% vs 96.7%) and accuracy (78.5% vs 69.6%) compared with zero-shot. Conclusion: RXL-RADSet provides a radiologist-verified multi-RADS benchmark; large SLMs (20-32B) can approach proprietary-model performance under guided prompting, but gaps remain for higher-complexity schemes.
Open Materials Generation with Stochastic Interpolants
Feb 04, 2025Abstract:The discovery of new materials is essential for enabling technological advancements. Computational approaches for predicting novel materials must effectively learn the manifold of stable crystal structures within an infinite design space. We introduce Open Materials Generation (OMG), a unifying framework for the generative design and discovery of inorganic crystalline materials. OMG employs stochastic interpolants (SI) to bridge an arbitrary base distribution to the target distribution of inorganic crystals via a broad class of tunable stochastic processes, encompassing both diffusion models and flow matching as special cases. In this work, we adapt the SI framework by integrating an equivariant graph representation of crystal structures and extending it to account for periodic boundary conditions in unit cell representations. Additionally, we couple the SI flow over spatial coordinates and lattice vectors with discrete flow matching for atomic species. We benchmark OMG's performance on two tasks: Crystal Structure Prediction (CSP) for specified compositions, and 'de novo' generation (DNG) aimed at discovering stable, novel, and unique structures. In our ground-up implementation of OMG, we refine and extend both CSP and DNG metrics compared to previous works. OMG establishes a new state-of-the-art in generative modeling for materials discovery, outperforming purely flow-based and diffusion-based implementations. These results underscore the importance of designing flexible deep learning frameworks to accelerate progress in materials science.
Novel Radiomic Measurements of Tumor- Associated Vasculature Morphology on Clinical Imaging as a Biomarker of Treatment Response in Multiple Cancers
Oct 05, 2022Abstract:Purpose: Tumor-associated vasculature differs from healthy blood vessels by its chaotic architecture and twistedness, which promotes treatment resistance. Measurable differences in these attributes may help stratify patients by likely benefit of systemic therapy (e.g. chemotherapy). In this work, we present a new category of radiomic biomarkers called quantitative tumor-associated vasculature (QuanTAV) features, and demonstrate their ability to predict response and survival across multiple cancers, imaging modalities, and treatment regimens. Experimental Design: We segmented tumor vessels and computed mathematical measurements of twistedness and organization on routine pre-treatment radiology (CT or contrast-enhanced MRI) from 558 patients, who received one of four first-line chemotherapy-based therapeutic intervention strategies for breast (n=371) or non-small cell lung cancer (NSCLC, n=187). Results: Across 4 chemotherapy-based treatment strategies, classifiers of QuanTAV measurements significantly (p<.05) predicted response in held out testing cohorts alone (AUC=0.63-0.71) and increased AUC by 0.06-0.12 when added to models of significant clinical variables alone. QuanTAV risk scores were prognostic of recurrence free survival in treatment cohorts chemotherapy for breast cancer (p=0.002, HR=1.25, 95% CI 1.08-1.44, C-index=.66) and chemoradiation for NSCLC (p=0.039, HR=1.28, 95% CI 1.01-1.62, C-index=0.66). Categorical QuanTAV risk groups were independently prognostic among all treatment groups, including NSCLC patients receiving chemotherapy (p=0.034, HR=2.29, 95% CI 1.07-4.94, C-index=0.62). Conclusions: Across these domains, we observed an association of vascular morphology on radiology with treatment outcome. Our findings suggest the potential of tumor-associated vasculature shape and structure as a prognostic and predictive biomarker for multiple cancers and treatments.
Lung Swapping Autoencoder: Learning a Disentangled Structure-texture Representation of Chest Radiographs
Jan 18, 2022


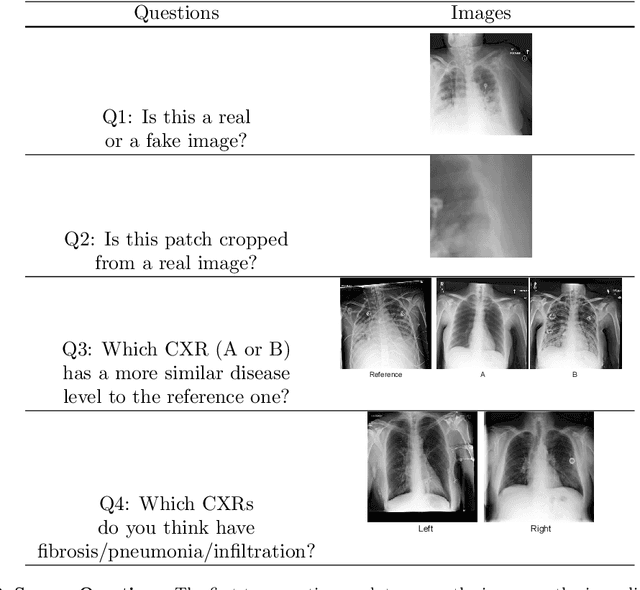
Abstract:Well-labeled datasets of chest radiographs (CXRs) are difficult to acquire due to the high cost of annotation. Thus, it is desirable to learn a robust and transferable representation in an unsupervised manner to benefit tasks that lack labeled data. Unlike natural images, medical images have their own domain prior; e.g., we observe that many pulmonary diseases, such as the COVID-19, manifest as changes in the lung tissue texture rather than the anatomical structure. Therefore, we hypothesize that studying only the texture without the influence of structure variations would be advantageous for downstream prognostic and predictive modeling tasks. In this paper, we propose a generative framework, the Lung Swapping Autoencoder (LSAE), that learns factorized representations of a CXR to disentangle the texture factor from the structure factor. Specifically, by adversarial training, the LSAE is optimized to generate a hybrid image that preserves the lung shape in one image but inherits the lung texture of another. To demonstrate the effectiveness of the disentangled texture representation, we evaluate the texture encoder $Enc^t$ in LSAE on ChestX-ray14 (N=112,120), and our own multi-institutional COVID-19 outcome prediction dataset, COVOC (N=340 (Subset-1) + 53 (Subset-2)). On both datasets, we reach or surpass the state-of-the-art by finetuning $Enc^t$ in LSAE that is 77% smaller than a baseline Inception v3. Additionally, in semi-and-self supervised settings with a similar model budget, $Enc^t$ in LSAE is also competitive with the state-of-the-art MoCo. By "re-mixing" the texture and shape factors, we generate meaningful hybrid images that can augment the training set. This data augmentation method can further improve COVOC prediction performance. The improvement is consistent even when we directly evaluate the Subset-1 trained model on Subset-2 without any fine-tuning.
Taxonomy Induction using Hypernym Subsequences
Sep 14, 2017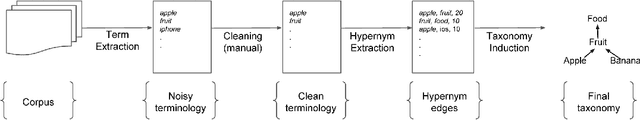
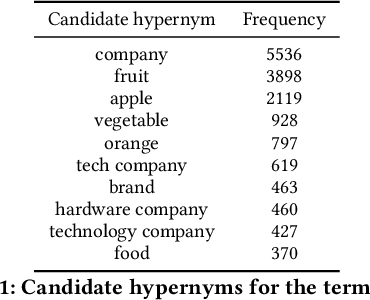
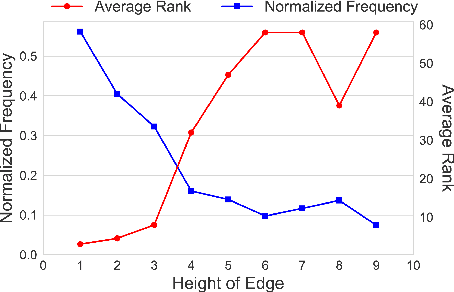
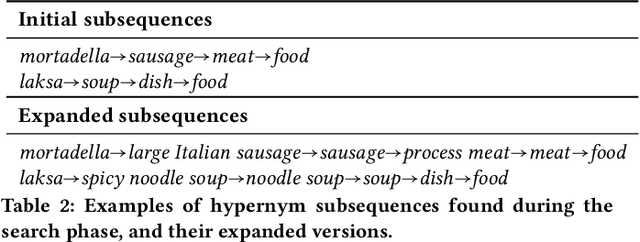
Abstract:We propose a novel, semi-supervised approach towards domain taxonomy induction from an input vocabulary of seed terms. Unlike all previous approaches, which typically extract direct hypernym edges for terms, our approach utilizes a novel probabilistic framework to extract hypernym subsequences. Taxonomy induction from extracted subsequences is cast as an instance of the minimumcost flow problem on a carefully designed directed graph. Through experiments, we demonstrate that our approach outperforms stateof- the-art taxonomy induction approaches across four languages. Importantly, we also show that our approach is robust to the presence of noise in the input vocabulary. To the best of our knowledge, no previous approaches have been empirically proven to manifest noise-robustness in the input vocabulary.
280 Birds with One Stone: Inducing Multilingual Taxonomies from Wikipedia using Character-level Classification
Sep 12, 2017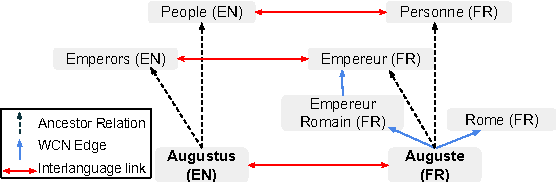

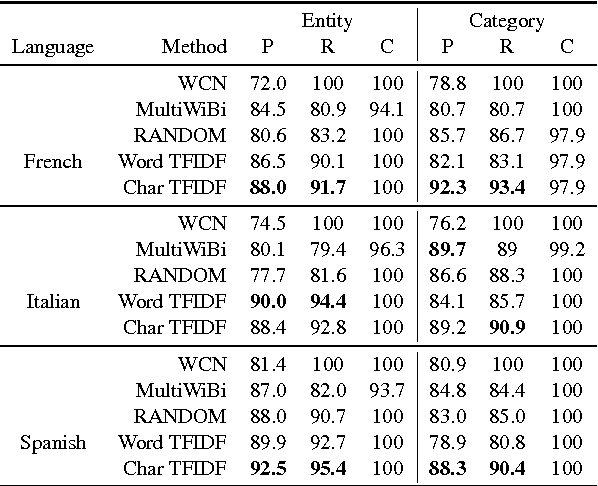
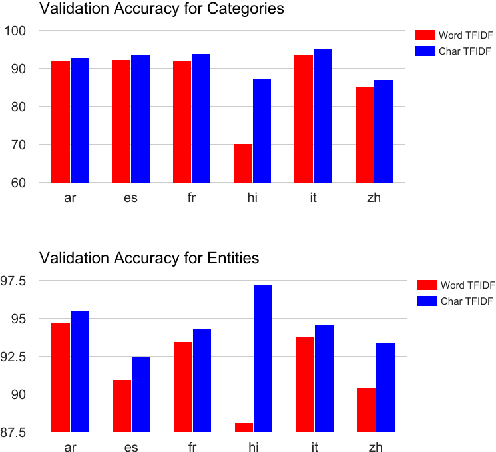
Abstract:We propose a simple, yet effective, approach towards inducing multilingual taxonomies from Wikipedia. Given an English taxonomy, our approach leverages the interlanguage links of Wikipedia followed by character-level classifiers to induce high-precision, high-coverage taxonomies in other languages. Through experiments, we demonstrate that our approach significantly outperforms the state-of-the-art, heuristics-heavy approaches for six languages. As a consequence of our work, we release presumably the largest and the most accurate multilingual taxonomic resource spanning over 280 languages.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge