Kaustav Bera
Novel Radiomic Measurements of Tumor- Associated Vasculature Morphology on Clinical Imaging as a Biomarker of Treatment Response in Multiple Cancers
Oct 05, 2022Abstract:Purpose: Tumor-associated vasculature differs from healthy blood vessels by its chaotic architecture and twistedness, which promotes treatment resistance. Measurable differences in these attributes may help stratify patients by likely benefit of systemic therapy (e.g. chemotherapy). In this work, we present a new category of radiomic biomarkers called quantitative tumor-associated vasculature (QuanTAV) features, and demonstrate their ability to predict response and survival across multiple cancers, imaging modalities, and treatment regimens. Experimental Design: We segmented tumor vessels and computed mathematical measurements of twistedness and organization on routine pre-treatment radiology (CT or contrast-enhanced MRI) from 558 patients, who received one of four first-line chemotherapy-based therapeutic intervention strategies for breast (n=371) or non-small cell lung cancer (NSCLC, n=187). Results: Across 4 chemotherapy-based treatment strategies, classifiers of QuanTAV measurements significantly (p<.05) predicted response in held out testing cohorts alone (AUC=0.63-0.71) and increased AUC by 0.06-0.12 when added to models of significant clinical variables alone. QuanTAV risk scores were prognostic of recurrence free survival in treatment cohorts chemotherapy for breast cancer (p=0.002, HR=1.25, 95% CI 1.08-1.44, C-index=.66) and chemoradiation for NSCLC (p=0.039, HR=1.28, 95% CI 1.01-1.62, C-index=0.66). Categorical QuanTAV risk groups were independently prognostic among all treatment groups, including NSCLC patients receiving chemotherapy (p=0.034, HR=2.29, 95% CI 1.07-4.94, C-index=0.62). Conclusions: Across these domains, we observed an association of vascular morphology on radiology with treatment outcome. Our findings suggest the potential of tumor-associated vasculature shape and structure as a prognostic and predictive biomarker for multiple cancers and treatments.
Radiomic Deformation and Textural Heterogeneity (R-DepTH) Descriptor to characterize Tumor Field Effect: Application to Survival Prediction in Glioblastoma
Mar 12, 2021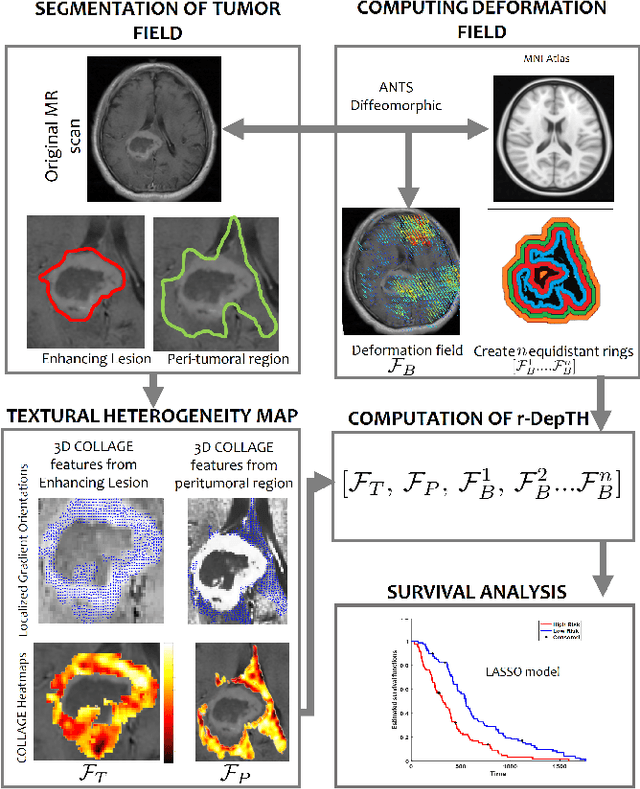
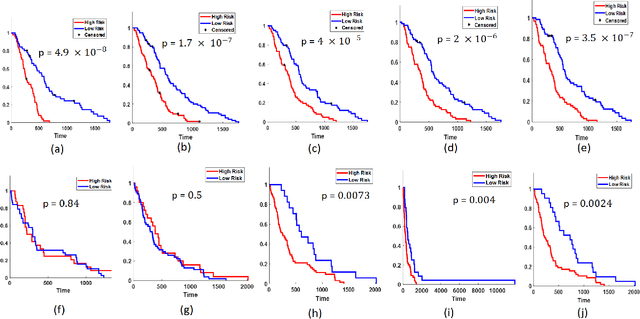
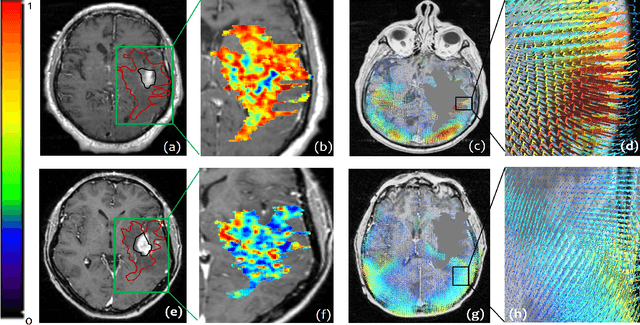
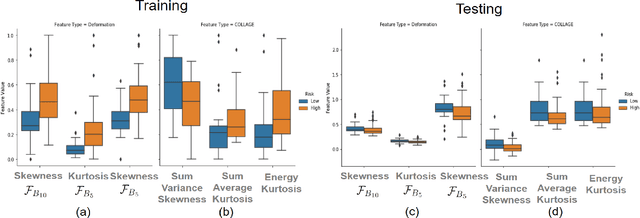
Abstract:The concept of tumor field effect implies that cancer is a systemic disease with its impact way beyond the visible tumor confines. For instance, in Glioblastoma (GBM), an aggressive brain tumor, the increase in intracranial pressure due to tumor burden often leads to brain herniation and poor outcomes. Our work is based on the rationale that highly aggressive tumors tend to grow uncontrollably, leading to pronounced biomechanical tissue deformations in the normal parenchyma, which when combined with local morphological differences in the tumor confines on MRI scans, will comprehensively capture tumor field effect. Specifically, we present an integrated MRI-based descriptor, radiomic-Deformation and Textural Heterogeneity (r-DepTH). This descriptor comprises measurements of the subtle perturbations in tissue deformations throughout the surrounding normal parenchyma due to mass effect. This involves non-rigidly aligning the patients MRI scans to a healthy atlas via diffeomorphic registration. The resulting inverse mapping is used to obtain the deformation field magnitudes in the normal parenchyma. These measurements are then combined with a 3D texture descriptor, Co-occurrence of Local Anisotropic Gradient Orientations (COLLAGE), which captures the morphological heterogeneity within the tumor confines, on MRI scans. R-DepTH, on N = 207 GBM cases (training set (St) = 128, testing set (Sv) = 79), demonstrated improved prognosis of overall survival by categorizing patients into low- (prolonged survival) and high-risk (poor survival) groups (on St, p-value = 0.0000035, and on Sv, p-value = 0.0024). R-DepTH descriptor may serve as a comprehensive MRI-based prognostic marker of disease aggressiveness and survival in solid tumors.
Spatial-And-Context aware (SpACe) "virtual biopsy" radiogenomic maps to target tumor mutational status on structural MRI
Jun 17, 2020
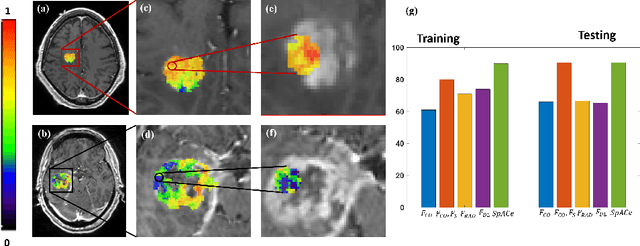
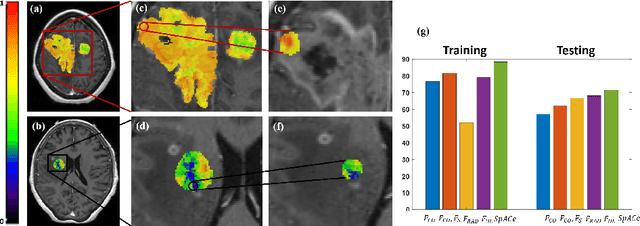
Abstract:With growing emphasis on personalized cancer-therapies,radiogenomics has shown promise in identifying target tumor mutational status on routine imaging (i.e. MRI) scans. These approaches fall into 2 categories: (1) deep-learning/radiomics (context-based), using image features from the entire tumor to identify the gene mutation status, or (2) atlas (spatial)-based to obtain likelihood of gene mutation status based on population statistics. While many genes (i.e. EGFR, MGMT) are spatially variant, a significant challenge in reliable assessment of gene mutation status on imaging has been the lack of available co-localized ground truth for training the models. We present Spatial-And-Context aware (SpACe) "virtual biopsy" maps that incorporate context-features from co-localized biopsy site along with spatial-priors from population atlases, within a Least Absolute Shrinkage and Selection Operator (LASSO) regression model, to obtain a per-voxel probability of the presence of a mutation status (M+ vs M-). We then use probabilistic pair-wise Markov model to improve the voxel-wise prediction probability. We evaluate the efficacy of SpACe maps on MRI scans with co-localized ground truth obtained from corresponding biopsy, to predict the mutation status of 2 driver genes in Glioblastoma: (1) EGFR (n=91), and (2) MGMT (n=81). When compared against deep-learning (DL) and radiomic models, SpACe maps obtained training and testing accuracies of 90% (n=71) and 90.48% (n=21) in identifying EGFR amplification status,compared to 80% and 71.4% via radiomics, and 74.28% and 65.5% via DL. For MGMT status, training and testing accuracies using SpACe were 88.3% (n=61) and 71.5% (n=20), compared to 52.4% and 66.7% using radiomics,and 79.3% and 68.4% using DL. Following validation,SpACe maps could provide surgical navigation to improve localization of sampling sites for targeting of specific driver genes in cancer.
Can tumor location on pre-treatment MRI predict likelihood of pseudo-progression versus tumor recurrence in Glioblastoma? A feasibility study
Jun 16, 2020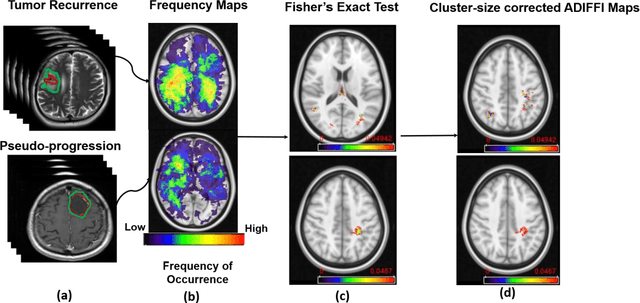

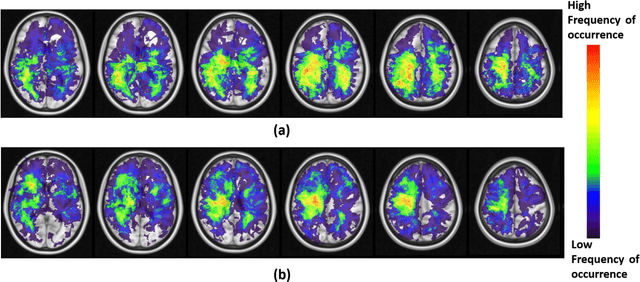
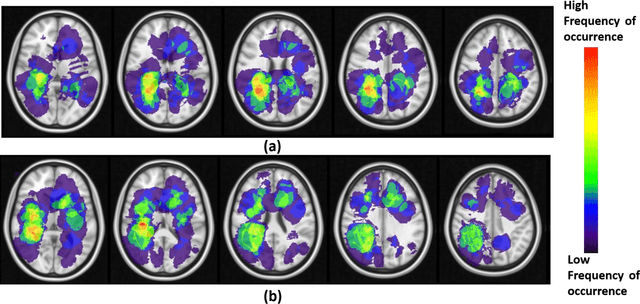
Abstract:A significant challenge in Glioblastoma (GBM) management is identifying pseudo-progression (PsP), a benign radiation-induced effect, from tumor recurrence, on routine imaging following conventional treatment. Previous studies have linked tumor lobar presence and laterality to GBM outcomes, suggesting that disease etiology and progression in GBM may be impacted by tumor location. Hence, in this feasibility study, we seek to investigate the following question: Can tumor location on treatment-na\"ive MRI provide early cues regarding likelihood of a patient developing pseudo-progression versus tumor recurrence? In this study, 74 pre-treatment Glioblastoma MRI scans with PsP (33) and tumor recurrence (41) were analyzed. First, enhancing lesion on Gd-T1w MRI and peri-lesional hyperintensities on T2w/FLAIR were segmented by experts and then registered to a brain atlas. Using patients from the two phenotypes, we construct two atlases by quantifying frequency of occurrence of enhancing lesion and peri-lesion hyperintensities, by averaging voxel intensities across the population. Analysis of differential involvement was then performed to compute voxel-wise significant differences (p-value<0.05) across the atlases. Statistically significant clusters were finally mapped to a structural atlas to provide anatomic localization of their location. Our results demonstrate that patients with tumor recurrence showed prominence of their initial tumor in the parietal lobe, while patients with PsP showed a multi-focal distribution of the initial tumor in the frontal and temporal lobes, insula, and putamen. These preliminary results suggest that lateralization of pre-treatment lesions towards certain anatomical areas of the brain may allow to provide early cues regarding assessing likelihood of occurrence of pseudo-progression from tumor recurrence on MRI scans.
Deep learning-based prediction of response to HER2-targeted neoadjuvant chemotherapy from pre-treatment dynamic breast MRI: A multi-institutional validation study
Jan 22, 2020



Abstract:Predicting response to neoadjuvant therapy is a vexing challenge in breast cancer. In this study, we evaluate the ability of deep learning to predict response to HER2-targeted neo-adjuvant chemotherapy (NAC) from pre-treatment dynamic contrast-enhanced (DCE) MRI acquired prior to treatment. In a retrospective study encompassing DCE-MRI data from a total of 157 HER2+ breast cancer patients from 5 institutions, we developed and validated a deep learning approach for predicting pathological complete response (pCR) to HER2-targeted NAC prior to treatment. 100 patients who received HER2-targeted neoadjuvant chemotherapy at a single institution were used to train (n=85) and tune (n=15) a convolutional neural network (CNN) to predict pCR. A multi-input CNN leveraging both pre-contrast and late post-contrast DCE-MRI acquisitions was identified to achieve optimal response prediction within the validation set (AUC=0.93). This model was then tested on two independent testing cohorts with pre-treatment DCE-MRI data. It achieved strong performance in a 28 patient testing set from a second institution (AUC=0.85, 95% CI 0.67-1.0, p=.0008) and a 29 patient multicenter trial including data from 3 additional institutions (AUC=0.77, 95% CI 0.58-0.97, p=0.006). Deep learning-based response prediction model was found to exceed a multivariable model incorporating predictive clinical variables (AUC < .65 in testing cohorts) and a model of semi-quantitative DCE-MRI pharmacokinetic measurements (AUC < .60 in testing cohorts). The results presented in this work across multiple sites suggest that with further validation deep learning could provide an effective and reliable tool to guide targeted therapy in breast cancer, thus reducing overtreatment among HER2+ patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge