Anant Madabhushi
Case Western Reserve University, Department of Biomedical Engineering, Cleveland OH, USA, Louis Stokes Veterans Administration Medical Center, Cleveland, OH, USA
SAGE: Agentic Framework for Interpretable and Clinically Translatable Computational Pathology Biomarker Discovery
Feb 01, 2026Abstract:Despite significant progress in computational pathology, many AI models remain black-box and difficult to interpret, posing a major barrier to clinical adoption due to limited transparency and explainability. This has motivated continued interest in engineered image-based biomarkers, which offer greater interpretability but are often proposed based on anecdotal evidence or fragmented prior literature rather than systematic biological validation. We introduce SAGE (Structured Agentic system for hypothesis Generation and Evaluation), an agentic AI system designed to identify interpretable, engineered pathology biomarkers by grounding them in biological evidence. SAGE integrates literature-anchored reasoning with multimodal data analysis to correlate image-derived features with molecular biomarkers, such as gene expression, and clinically relevant outcomes. By coordinating specialized agents for biological contextualization and empirical hypothesis validation, SAGE prioritizes transparent, biologically supported biomarkers and advances the clinical translation of computational pathology.
UNetVL: Enhancing 3D Medical Image Segmentation with Chebyshev KAN Powered Vision-LSTM
Jan 13, 2025



Abstract:3D medical image segmentation has progressed considerably due to Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs), yet these methods struggle to balance long-range dependency acquisition with computational efficiency. To address this challenge, we propose UNETVL (U-Net Vision-LSTM), a novel architecture that leverages recent advancements in temporal information processing. UNETVL incorporates Vision-LSTM (ViL) for improved scalability and memory functions, alongside an efficient Chebyshev Kolmogorov-Arnold Networks (KAN) to handle complex and long-range dependency patterns more effectively. We validated our method on the ACDC and AMOS2022 (post challenge Task 2) benchmark datasets, showing a significant improvement in mean Dice score compared to recent state-of-the-art approaches, especially over its predecessor, UNETR, with increases of 7.3% on ACDC and 15.6% on AMOS, respectively. Extensive ablation studies were conducted to demonstrate the impact of each component in UNETVL, providing a comprehensive understanding of its architecture. Our code is available at https://github.com/tgrex6/UNETVL, facilitating further research and applications in this domain.
CohortFinder: an open-source tool for data-driven partitioning of biomedical image cohorts to yield robust machine learning models
Jul 17, 2023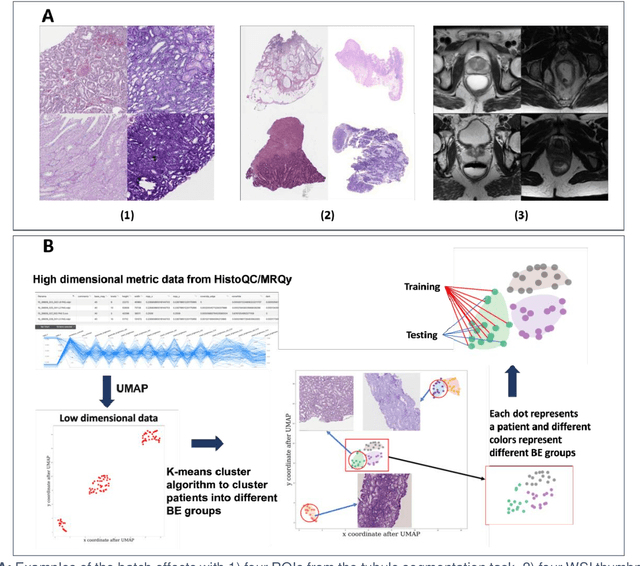

Abstract:Batch effects (BEs) refer to systematic technical differences in data collection unrelated to biological variations whose noise is shown to negatively impact machine learning (ML) model generalizability. Here we release CohortFinder, an open-source tool aimed at mitigating BEs via data-driven cohort partitioning. We demonstrate CohortFinder improves ML model performance in downstream medical image processing tasks. CohortFinder is freely available for download at cohortfinder.com.
PatchSorter: A High Throughput Deep Learning Digital Pathology Tool for Object Labeling
Jul 13, 2023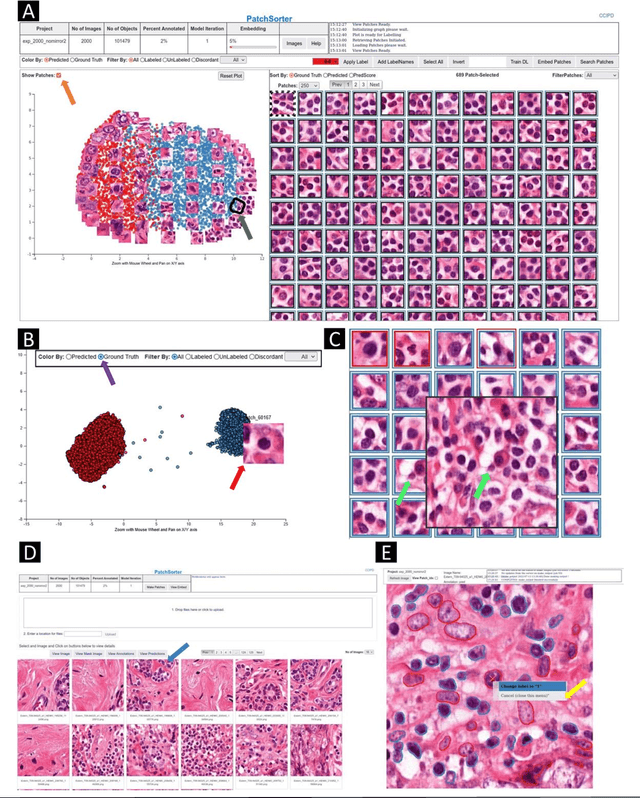
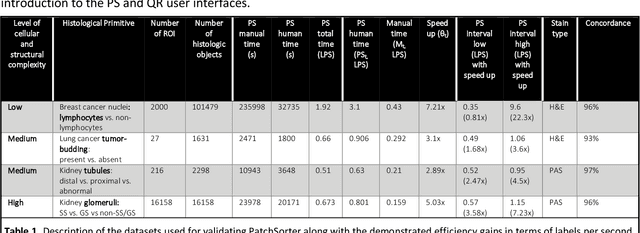
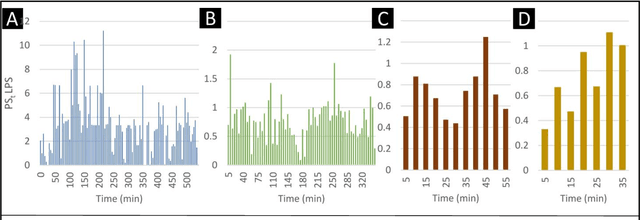
Abstract:The discovery of patterns associated with diagnosis, prognosis, and therapy response in digital pathology images often requires intractable labeling of large quantities of histological objects. Here we release an open-source labeling tool, PatchSorter, which integrates deep learning with an intuitive web interface. Using >100,000 objects, we demonstrate a >7x improvement in labels per second over unaided labeling, with minimal impact on labeling accuracy, thus enabling high-throughput labeling of large datasets.
Breast Cancer Immunohistochemical Image Generation: a Benchmark Dataset and Challenge Review
May 05, 2023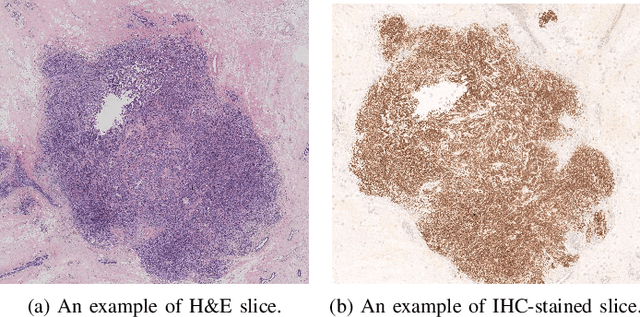
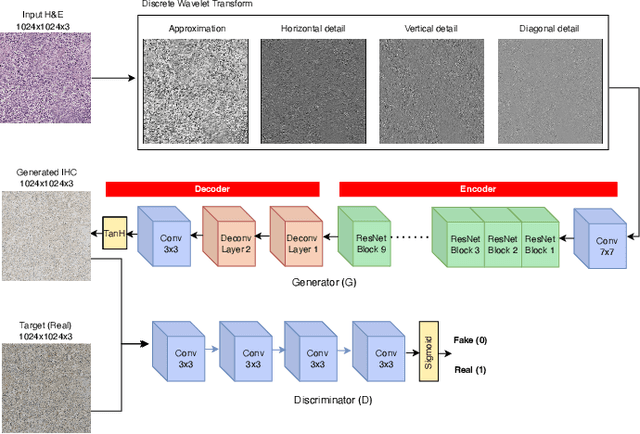
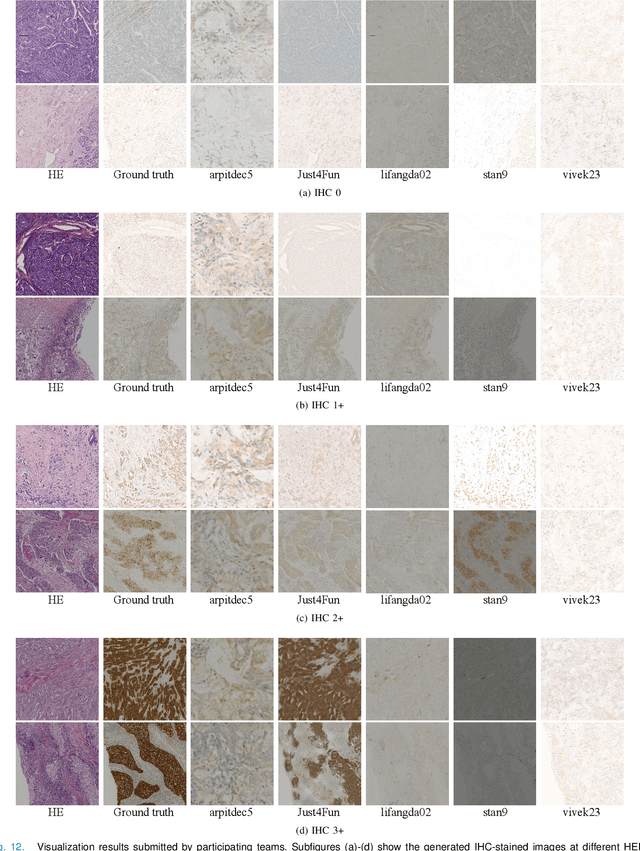
Abstract:For invasive breast cancer, immunohistochemical (IHC) techniques are often used to detect the expression level of human epidermal growth factor receptor-2 (HER2) in breast tissue to formulate a precise treatment plan. From the perspective of saving manpower, material and time costs, directly generating IHC-stained images from hematoxylin and eosin (H&E) stained images is a valuable research direction. Therefore, we held the breast cancer immunohistochemical image generation challenge, aiming to explore novel ideas of deep learning technology in pathological image generation and promote research in this field. The challenge provided registered H&E and IHC-stained image pairs, and participants were required to use these images to train a model that can directly generate IHC-stained images from corresponding H&E-stained images. We selected and reviewed the five highest-ranking methods based on their PSNR and SSIM metrics, while also providing overviews of the corresponding pipelines and implementations. In this paper, we further analyze the current limitations in the field of breast cancer immunohistochemical image generation and forecast the future development of this field. We hope that the released dataset and the challenge will inspire more scholars to jointly study higher-quality IHC-stained image generation.
Novel Radiomic Measurements of Tumor- Associated Vasculature Morphology on Clinical Imaging as a Biomarker of Treatment Response in Multiple Cancers
Oct 05, 2022Abstract:Purpose: Tumor-associated vasculature differs from healthy blood vessels by its chaotic architecture and twistedness, which promotes treatment resistance. Measurable differences in these attributes may help stratify patients by likely benefit of systemic therapy (e.g. chemotherapy). In this work, we present a new category of radiomic biomarkers called quantitative tumor-associated vasculature (QuanTAV) features, and demonstrate their ability to predict response and survival across multiple cancers, imaging modalities, and treatment regimens. Experimental Design: We segmented tumor vessels and computed mathematical measurements of twistedness and organization on routine pre-treatment radiology (CT or contrast-enhanced MRI) from 558 patients, who received one of four first-line chemotherapy-based therapeutic intervention strategies for breast (n=371) or non-small cell lung cancer (NSCLC, n=187). Results: Across 4 chemotherapy-based treatment strategies, classifiers of QuanTAV measurements significantly (p<.05) predicted response in held out testing cohorts alone (AUC=0.63-0.71) and increased AUC by 0.06-0.12 when added to models of significant clinical variables alone. QuanTAV risk scores were prognostic of recurrence free survival in treatment cohorts chemotherapy for breast cancer (p=0.002, HR=1.25, 95% CI 1.08-1.44, C-index=.66) and chemoradiation for NSCLC (p=0.039, HR=1.28, 95% CI 1.01-1.62, C-index=0.66). Categorical QuanTAV risk groups were independently prognostic among all treatment groups, including NSCLC patients receiving chemotherapy (p=0.034, HR=2.29, 95% CI 1.07-4.94, C-index=0.62). Conclusions: Across these domains, we observed an association of vascular morphology on radiology with treatment outcome. Our findings suggest the potential of tumor-associated vasculature shape and structure as a prognostic and predictive biomarker for multiple cancers and treatments.
Radiomic Deformation and Textural Heterogeneity (R-DepTH) Descriptor to characterize Tumor Field Effect: Application to Survival Prediction in Glioblastoma
Mar 12, 2021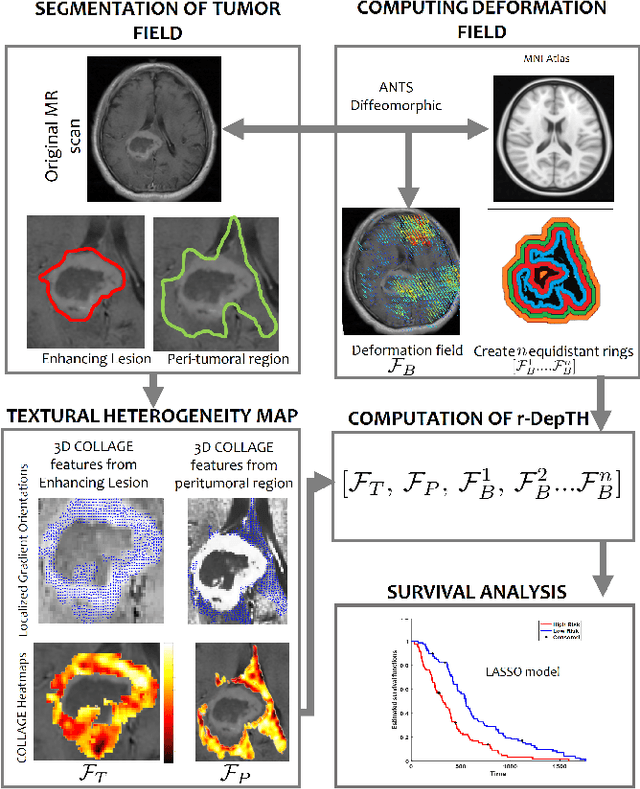
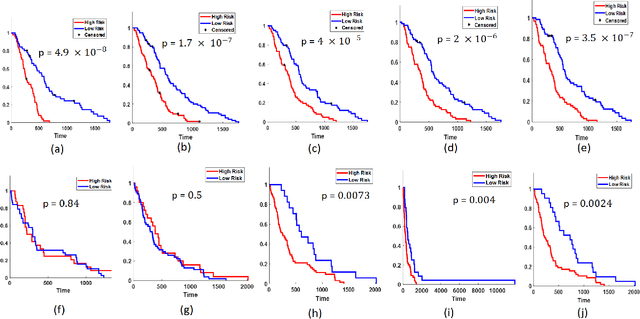
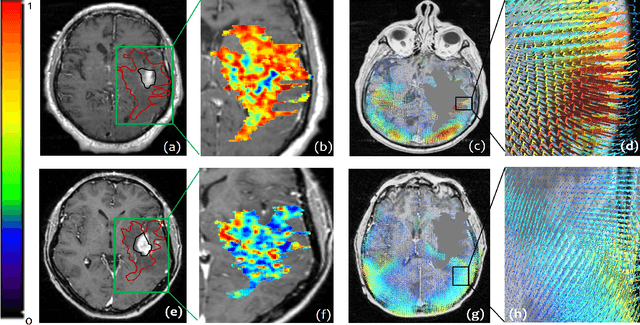
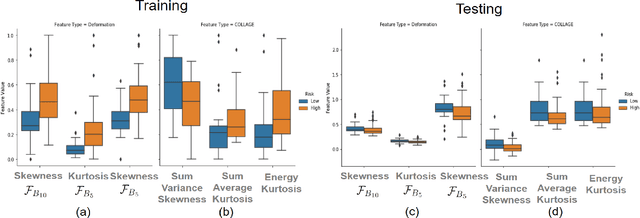
Abstract:The concept of tumor field effect implies that cancer is a systemic disease with its impact way beyond the visible tumor confines. For instance, in Glioblastoma (GBM), an aggressive brain tumor, the increase in intracranial pressure due to tumor burden often leads to brain herniation and poor outcomes. Our work is based on the rationale that highly aggressive tumors tend to grow uncontrollably, leading to pronounced biomechanical tissue deformations in the normal parenchyma, which when combined with local morphological differences in the tumor confines on MRI scans, will comprehensively capture tumor field effect. Specifically, we present an integrated MRI-based descriptor, radiomic-Deformation and Textural Heterogeneity (r-DepTH). This descriptor comprises measurements of the subtle perturbations in tissue deformations throughout the surrounding normal parenchyma due to mass effect. This involves non-rigidly aligning the patients MRI scans to a healthy atlas via diffeomorphic registration. The resulting inverse mapping is used to obtain the deformation field magnitudes in the normal parenchyma. These measurements are then combined with a 3D texture descriptor, Co-occurrence of Local Anisotropic Gradient Orientations (COLLAGE), which captures the morphological heterogeneity within the tumor confines, on MRI scans. R-DepTH, on N = 207 GBM cases (training set (St) = 128, testing set (Sv) = 79), demonstrated improved prognosis of overall survival by categorizing patients into low- (prolonged survival) and high-risk (poor survival) groups (on St, p-value = 0.0000035, and on Sv, p-value = 0.0024). R-DepTH descriptor may serve as a comprehensive MRI-based prognostic marker of disease aggressiveness and survival in solid tumors.
Quick Annotator: an open-source digital pathology based rapid image annotation tool
Jan 06, 2021



Abstract:Image based biomarker discovery typically requires an accurate segmentation of histologic structures (e.g., cell nuclei, tubules, epithelial regions) in digital pathology Whole Slide Images (WSI). Unfortunately, annotating each structure of interest is laborious and often intractable even in moderately sized cohorts. Here, we present an open-source tool, Quick Annotator (QA), designed to improve annotation efficiency of histologic structures by orders of magnitude. While the user annotates regions of interest (ROI) via an intuitive web interface, a deep learning (DL) model is concurrently optimized using these annotations and applied to the ROI. The user iteratively reviews DL results to either (a) accept accurately annotated regions, or (b) correct erroneously segmented structures to improve subsequent model suggestions, before transitioning to other ROIs. We demonstrate the effectiveness of QA over comparable manual efforts via three use cases. These include annotating (a) 337,386 nuclei in 5 pancreatic WSIs, (b) 5,692 tubules in 10 colorectal WSIs, and (c) 14,187 regions of epithelium in 10 breast WSIs. Efficiency gains in terms of annotations per second of 102x, 9x, and 39x were respectively witnessed while retaining f-scores >.95, suggesting QA may be a valuable tool for efficiently fully annotating WSIs employed in downstream biomarker studies.
A review of deep learning in medical imaging: Image traits, technology trends, case studies with progress highlights, and future promises
Aug 02, 2020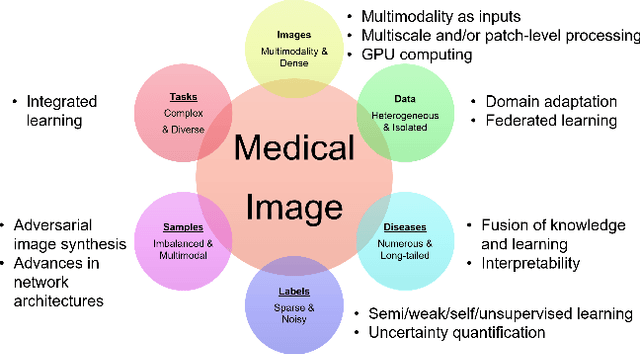
Abstract:Since its renaissance, deep learning has been widely used in various medical imaging tasks and has achieved remarkable success in many medical imaging applications, thereby propelling us into the so-called artificial intelligence (AI) era. It is known that the success of AI is mostly attributed to the availability of big data with annotations for a single task and the advances in high performance computing. However, medical imaging presents unique challenges that confront deep learning approaches. In this survey paper, we first highlight both clinical needs and technical challenges in medical imaging and describe how emerging trends in deep learning are addressing these issues. We cover the topics of network architecture, sparse and noisy labels, federating learning, interpretability, uncertainty quantification, etc. Then, we present several case studies that are commonly found in clinical practice, including digital pathology and chest, brain, cardiovascular, and abdominal imaging. Rather than presenting an exhaustive literature survey, we instead describe some prominent research highlights related to these case study applications. We conclude with a discussion and presentation of promising future directions.
Can tumor location on pre-treatment MRI predict likelihood of pseudo-progression versus tumor recurrence in Glioblastoma? A feasibility study
Jun 16, 2020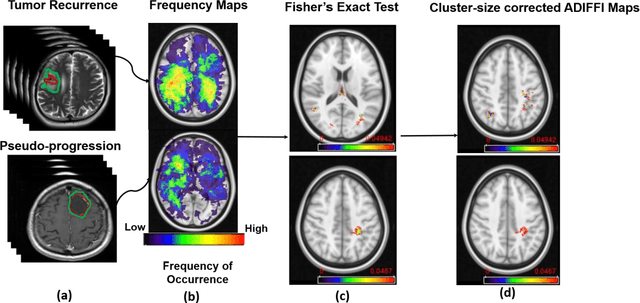

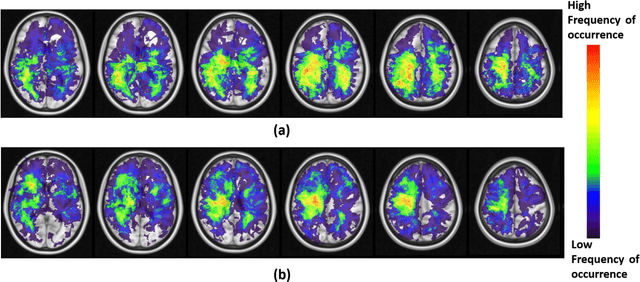
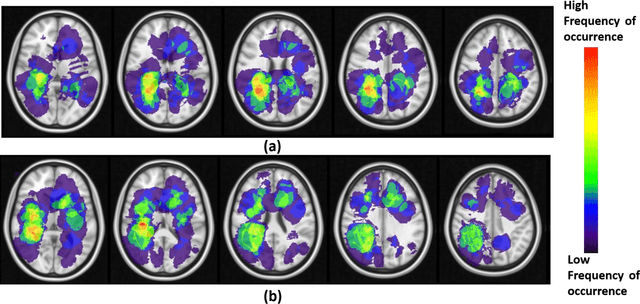
Abstract:A significant challenge in Glioblastoma (GBM) management is identifying pseudo-progression (PsP), a benign radiation-induced effect, from tumor recurrence, on routine imaging following conventional treatment. Previous studies have linked tumor lobar presence and laterality to GBM outcomes, suggesting that disease etiology and progression in GBM may be impacted by tumor location. Hence, in this feasibility study, we seek to investigate the following question: Can tumor location on treatment-na\"ive MRI provide early cues regarding likelihood of a patient developing pseudo-progression versus tumor recurrence? In this study, 74 pre-treatment Glioblastoma MRI scans with PsP (33) and tumor recurrence (41) were analyzed. First, enhancing lesion on Gd-T1w MRI and peri-lesional hyperintensities on T2w/FLAIR were segmented by experts and then registered to a brain atlas. Using patients from the two phenotypes, we construct two atlases by quantifying frequency of occurrence of enhancing lesion and peri-lesion hyperintensities, by averaging voxel intensities across the population. Analysis of differential involvement was then performed to compute voxel-wise significant differences (p-value<0.05) across the atlases. Statistically significant clusters were finally mapped to a structural atlas to provide anatomic localization of their location. Our results demonstrate that patients with tumor recurrence showed prominence of their initial tumor in the parietal lobe, while patients with PsP showed a multi-focal distribution of the initial tumor in the frontal and temporal lobes, insula, and putamen. These preliminary results suggest that lateralization of pre-treatment lesions towards certain anatomical areas of the brain may allow to provide early cues regarding assessing likelihood of occurrence of pseudo-progression from tumor recurrence on MRI scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge