Hayit Greenspan
ProtoSAM -- One Shot Medical Image Segmentation With Foundational Models
Jul 09, 2024



Abstract:This work introduces a new framework, ProtoSAM, for one-shot medical image segmentation. It combines the use of prototypical networks, known for few-shot segmentation, with SAM - a natural image foundation model. The method proposed creates an initial coarse segmentation mask using the ALPnet prototypical network, augmented with a DINOv2 encoder. Following the extraction of an initial mask, prompts are extracted, such as points and bounding boxes, which are then input into the Segment Anything Model (SAM). State-of-the-art results are shown on several medical image datasets and demonstrate automated segmentation capabilities using a single image example (one shot) with no need for fine-tuning of the foundation model.
X-ray2CTPA: Generating 3D CTPA scans from 2D X-ray conditioning
Jun 25, 2024Abstract:Chest X-rays or chest radiography (CXR), commonly used for medical diagnostics, typically enables limited imaging compared to computed tomography (CT) scans, which offer more detailed and accurate three-dimensional data, particularly contrast-enhanced scans like CT Pulmonary Angiography (CTPA). However, CT scans entail higher costs, greater radiation exposure, and are less accessible than CXRs. In this work we explore cross-modal translation from a 2D low contrast-resolution X-ray input to a 3D high contrast and spatial-resolution CTPA scan. Driven by recent advances in generative AI, we introduce a novel diffusion-based approach to this task. We evaluate the models performance using both quantitative metrics and qualitative feedback from radiologists, ensuring diagnostic relevance of the generated images. Furthermore, we employ the synthesized 3D images in a classification framework and show improved AUC in a PE categorization task, using the initial CXR input. The proposed method is generalizable and capable of performing additional cross-modality translations in medical imaging. It may pave the way for more accessible and cost-effective advanced diagnostic tools. The code for this project is available: https://github.com/NoaCahan/X-ray2CTPA .
DINOv2 based Self Supervised Learning For Few Shot Medical Image Segmentation
Mar 05, 2024



Abstract:Deep learning models have emerged as the cornerstone of medical image segmentation, but their efficacy hinges on the availability of extensive manually labeled datasets and their adaptability to unforeseen categories remains a challenge. Few-shot segmentation (FSS) offers a promising solution by endowing models with the capacity to learn novel classes from limited labeled examples. A leading method for FSS is ALPNet, which compares features between the query image and the few available support segmented images. A key question about using ALPNet is how to design its features. In this work, we delve into the potential of using features from DINOv2, which is a foundational self-supervised learning model in computer vision. Leveraging the strengths of ALPNet and harnessing the feature extraction capabilities of DINOv2, we present a novel approach to few-shot segmentation that not only enhances performance but also paves the way for more robust and adaptable medical image analysis.
A Self Supervised StyleGAN for Image Annotation and Classification with Extremely Limited Labels
Dec 26, 2023



Abstract:The recent success of learning-based algorithms can be greatly attributed to the immense amount of annotated data used for training. Yet, many datasets lack annotations due to the high costs associated with labeling, resulting in degraded performances of deep learning methods. Self-supervised learning is frequently adopted to mitigate the reliance on massive labeled datasets since it exploits unlabeled data to learn relevant feature representations. In this work, we propose SS-StyleGAN, a self-supervised approach for image annotation and classification suitable for extremely small annotated datasets. This novel framework adds self-supervision to the StyleGAN architecture by integrating an encoder that learns the embedding to the StyleGAN latent space, which is well-known for its disentangled properties. The learned latent space enables the smart selection of representatives from the data to be labeled for improved classification performance. We show that the proposed method attains strong classification results using small labeled datasets of sizes 50 and even 10. We demonstrate the superiority of our approach for the tasks of COVID-19 and liver tumor pathology identification.
* Accepted to IEEE Transactions on Medical Imaging
Neural Network Reconstruction of the Left Atrium using Sparse Catheter Paths
Nov 04, 2023Abstract:Catheter based radiofrequency ablation for pulmonary vein isolation has become the first line of treatment for atrial fibrillation in recent years. This requires a rather accurate map of the left atrial sub-endocardial surface including the ostia of the pulmonary veins, which requires dense sampling of the surface and takes more than 10 minutes. The focus of this work is to provide left atrial visualization early in the procedure to ease procedure complexity and enable further workflows, such as using catheters that have difficulty sampling the surface. We propose a dense encoder-decoder network with a novel regularization term to reconstruct the shape of the left atrium from partial data which is derived from simple catheter maneuvers. To train the network, we acquire a large dataset of 3D atria shapes and generate corresponding catheter trajectories. Once trained, we show that the suggested network can sufficiently approximate the atrium shape based on a given trajectory. We compare several network solutions for the 3D atrium reconstruction. We demonstrate that the solution proposed produces realistic visualization using partial acquisition within a 3-minute time interval. Synthetic and human clinical cases are shown.
Weakly Supervised Attention Model for RV StrainClassification from volumetric CTPA Scans
Jul 26, 2021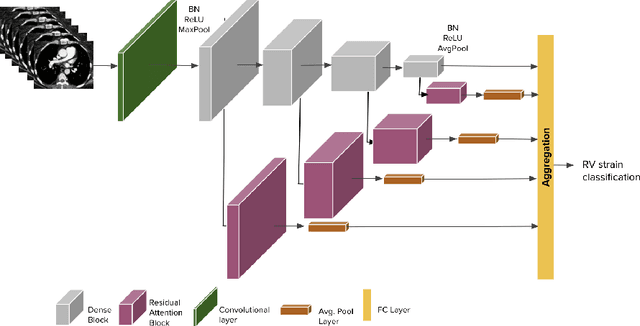
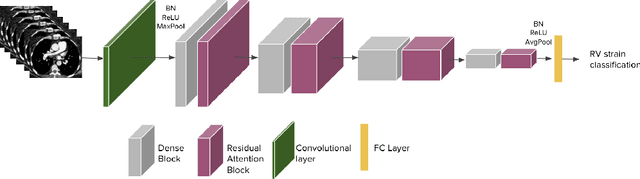
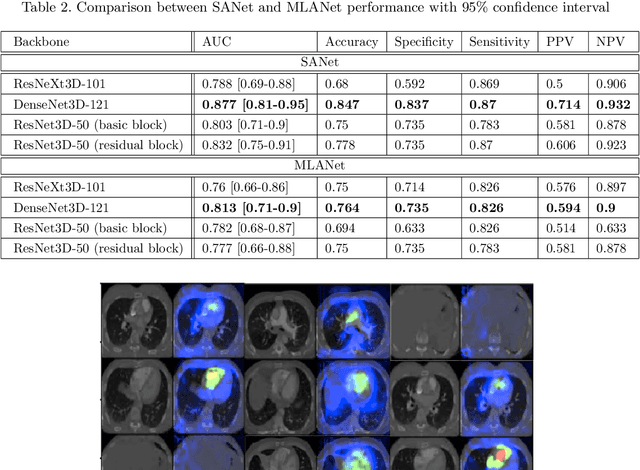
Abstract:Pulmonary embolus (PE) refers to obstruction of pulmonary arteries by blood clots. PE accounts for approximately 100,000 deaths per year in the United States alone. The clinical presentation of PE is often nonspecific, making the diagnosis challenging. Thus, rapid and accurate risk stratification is of paramount importance. High-risk PE is caused by right ventricular (RV) dysfunction from acute pressure overload, which in return can help identify which patients require more aggressive therapy. Reconstructed four-chamber views of the heart on chest CT can detect right ventricular enlargement. CT pulmonary angiography (CTPA) is the golden standard in the diagnostic workup of suspected PE. Therefore, it can link between diagnosis and risk stratification strategies. We developed a weakly supervised deep learning algorithm, with an emphasis on a novel attention mechanism, to automatically classify RV strain on CTPA. Our method is a 3D DenseNet model with integrated 3D residual attention blocks. We evaluated our model on a dataset of CTPAs of emergency department (ED) PE patients. This model achieved an area under the receiver operating characteristic curve (AUC) of 0.88 for classifying RV strain. The model showed a sensitivity of 87% and specificity of 83.7%. Our solution outperforms state-of-the-art 3D CNN networks. The proposed design allows for a fully automated network that can be trained easily in an end-to-end manner without requiring computationally intensive and time-consuming preprocessing or strenuous labeling of the data.We infer that unmarked CTPAs can be used for effective RV strain classification. This could be used as a second reader, alerting for high-risk PE patients. To the best of our knowledge, there are no previous deep learning-based studies that attempted to solve this problem.
Learning Rotation Invariant Features for Cryogenic Electron Microscopy Image Reconstruction
Jan 10, 2021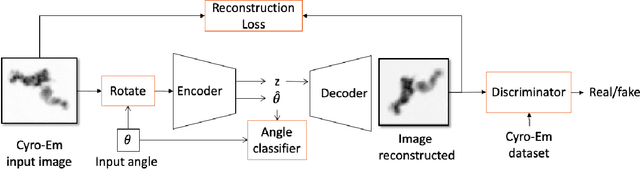
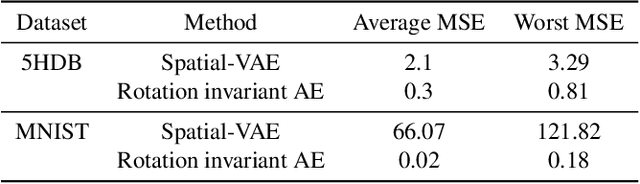
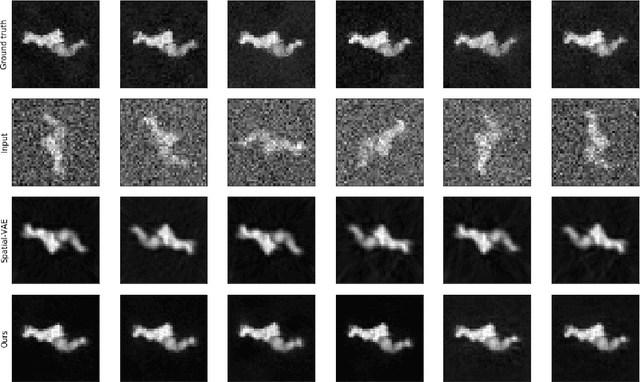
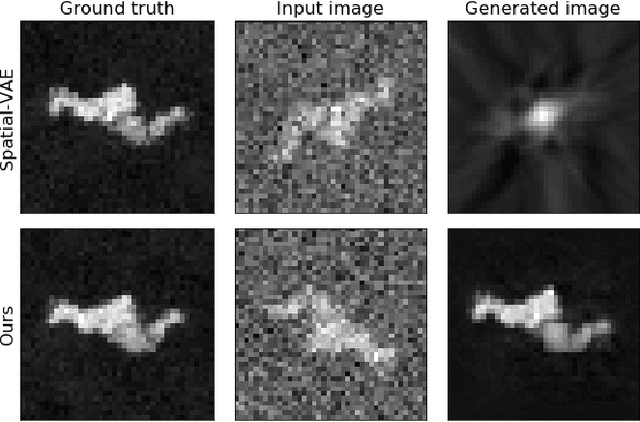
Abstract:Cryo-Electron Microscopy (Cryo-EM) is a Nobel prize-winning technology for determining the 3D structure of particles at near-atomic resolution. A fundamental step in the recovering of the 3D single-particle structure is to align its 2D projections; thus, the construction of a canonical representation with a fixed rotation angle is required. Most approaches use discrete clustering which fails to capture the continuous nature of image rotation, others suffer from low-quality image reconstruction. We propose a novel method that leverages the recent development in the generative adversarial networks. We introduce an encoder-decoder with a rotation angle classifier. In addition, we utilize a discriminator on the decoder output to minimize the reconstruction error. We demonstrate our approach with the Cryo-EM 5HDB and the rotated MNIST datasets showing substantial improvement over recent methods.
Automated triage of COVID-19 from various lung abnormalities using chest CT features
Oct 24, 2020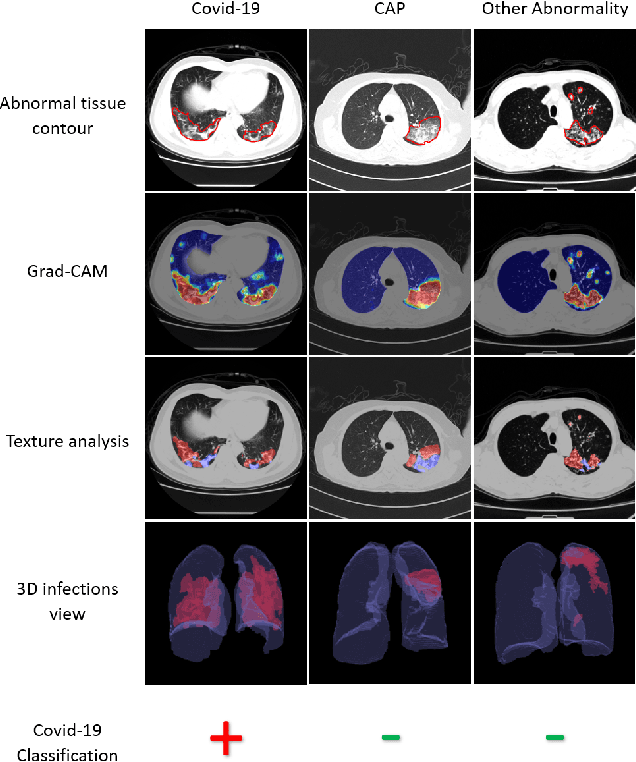

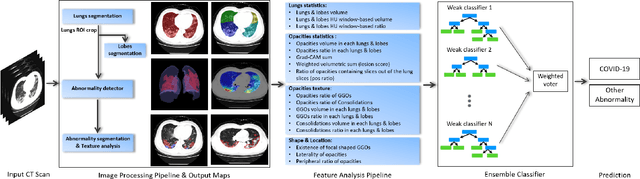
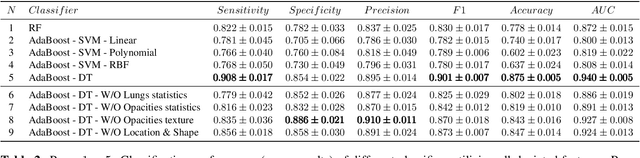
Abstract:The outbreak of COVID-19 has lead to a global effort to decelerate the pandemic spread. For this purpose chest computed-tomography (CT) based screening and diagnosis of COVID-19 suspected patients is utilized, either as a support or replacement to reverse transcription-polymerase chain reaction (RT-PCR) test. In this paper, we propose a fully automated AI based system that takes as input chest CT scans and triages COVID-19 cases. More specifically, we produce multiple descriptive features, including lung and infections statistics, texture, shape and location, to train a machine learning based classifier that distinguishes between COVID-19 and other lung abnormalities (including community acquired pneumonia). We evaluated our system on a dataset of 2191 CT cases and demonstrated a robust solution with 90.8% sensitivity at 85.4% specificity with 94.0% ROC-AUC. In addition, we present an elaborated feature analysis and ablation study to explore the importance of each feature.
COVID-19 in CXR: from Detection and Severity Scoring to Patient Disease Monitoring
Aug 04, 2020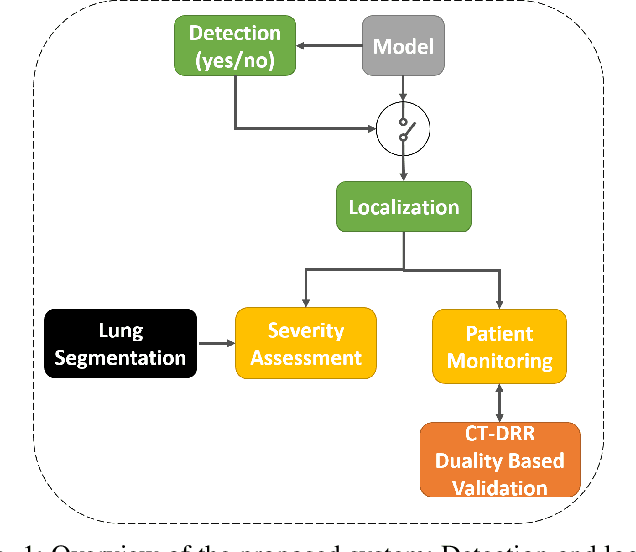
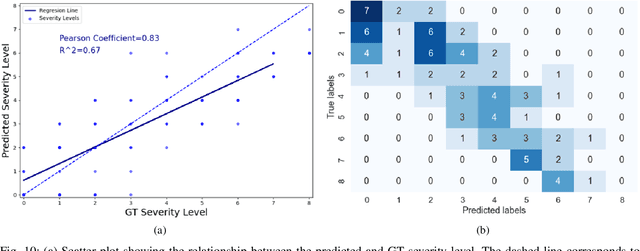
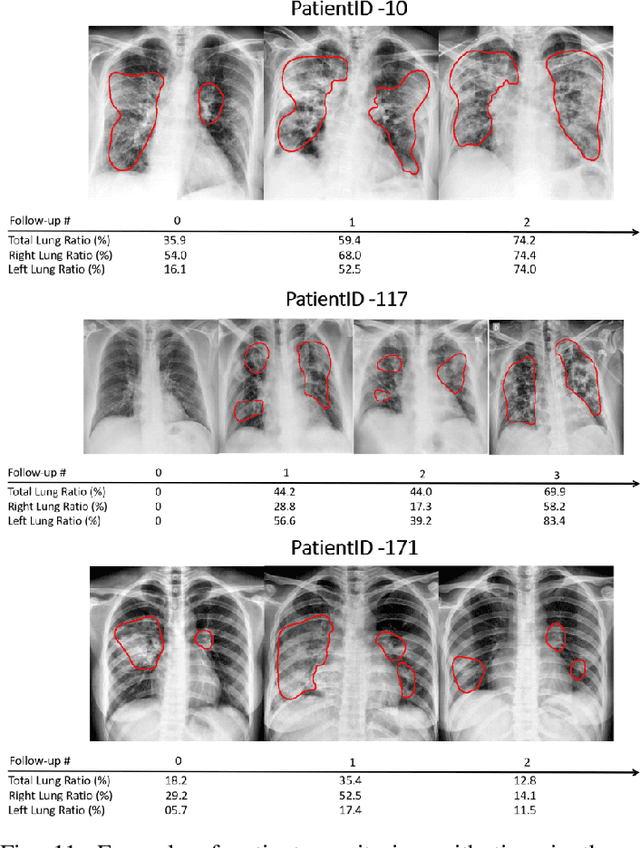
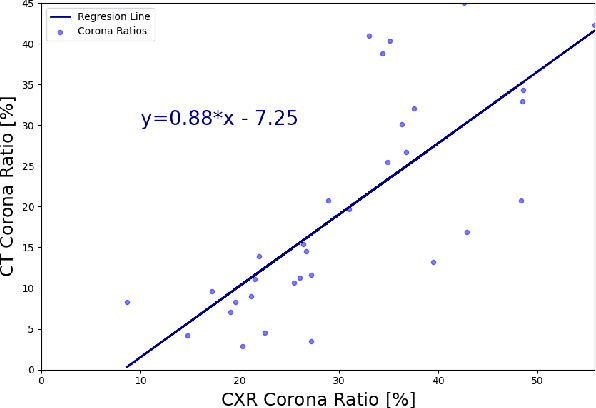
Abstract:In this work, we estimate the severity of pneumonia in COVID-19 patients and conduct a longitudinal study of disease progression. To achieve this goal, we developed a deep learning model for simultaneous detection and segmentation of pneumonia in chest Xray (CXR) images and generalized to COVID-19 pneumonia. The segmentations were utilized to calculate a "Pneumonia Ratio" which indicates the disease severity. The measurement of disease severity enables to build a disease extent profile over time for hospitalized patients. To validate the model relevance to the patient monitoring task, we developed a validation strategy which involves a synthesis of Digital Reconstructed Radiographs (DRRs - synthetic Xray) from serial CT scans; we then compared the disease progression profiles that were generated from the DRRs to those that were generated from CT volumes.
A review of deep learning in medical imaging: Image traits, technology trends, case studies with progress highlights, and future promises
Aug 02, 2020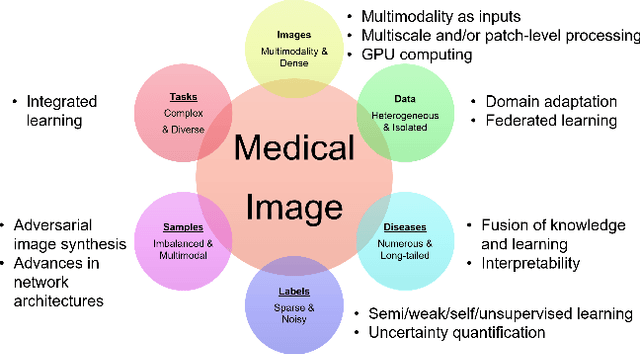
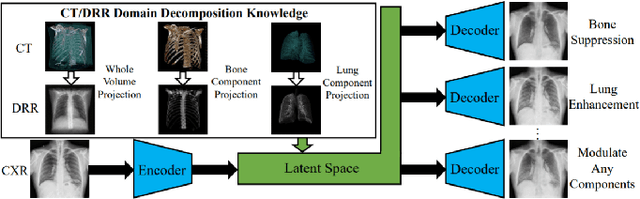
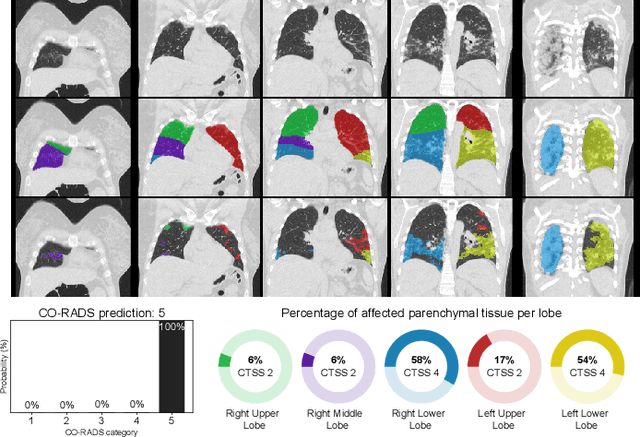
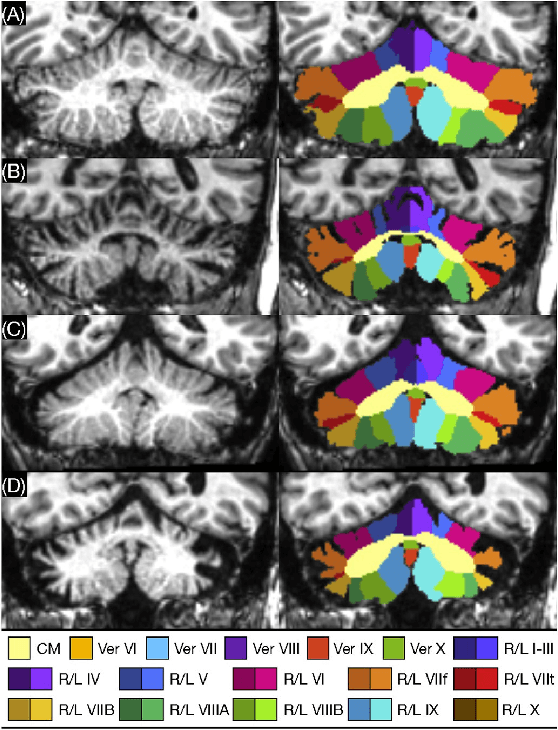
Abstract:Since its renaissance, deep learning has been widely used in various medical imaging tasks and has achieved remarkable success in many medical imaging applications, thereby propelling us into the so-called artificial intelligence (AI) era. It is known that the success of AI is mostly attributed to the availability of big data with annotations for a single task and the advances in high performance computing. However, medical imaging presents unique challenges that confront deep learning approaches. In this survey paper, we first highlight both clinical needs and technical challenges in medical imaging and describe how emerging trends in deep learning are addressing these issues. We cover the topics of network architecture, sparse and noisy labels, federating learning, interpretability, uncertainty quantification, etc. Then, we present several case studies that are commonly found in clinical practice, including digital pathology and chest, brain, cardiovascular, and abdominal imaging. Rather than presenting an exhaustive literature survey, we instead describe some prominent research highlights related to these case study applications. We conclude with a discussion and presentation of promising future directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge