Noa Cahan
X-ray2CTPA: Generating 3D CTPA scans from 2D X-ray conditioning
Jun 25, 2024Abstract:Chest X-rays or chest radiography (CXR), commonly used for medical diagnostics, typically enables limited imaging compared to computed tomography (CT) scans, which offer more detailed and accurate three-dimensional data, particularly contrast-enhanced scans like CT Pulmonary Angiography (CTPA). However, CT scans entail higher costs, greater radiation exposure, and are less accessible than CXRs. In this work we explore cross-modal translation from a 2D low contrast-resolution X-ray input to a 3D high contrast and spatial-resolution CTPA scan. Driven by recent advances in generative AI, we introduce a novel diffusion-based approach to this task. We evaluate the models performance using both quantitative metrics and qualitative feedback from radiologists, ensuring diagnostic relevance of the generated images. Furthermore, we employ the synthesized 3D images in a classification framework and show improved AUC in a PE categorization task, using the initial CXR input. The proposed method is generalizable and capable of performing additional cross-modality translations in medical imaging. It may pave the way for more accessible and cost-effective advanced diagnostic tools. The code for this project is available: https://github.com/NoaCahan/X-ray2CTPA .
Weakly Supervised Attention Model for RV StrainClassification from volumetric CTPA Scans
Jul 26, 2021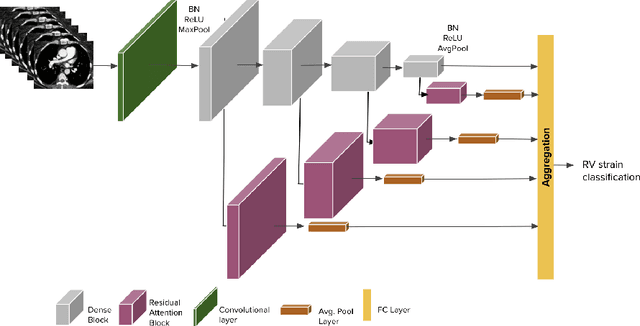
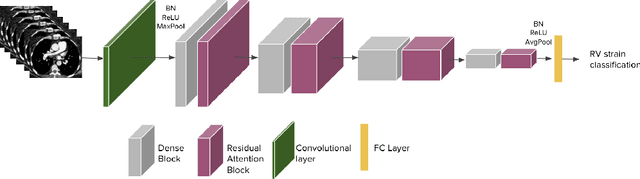
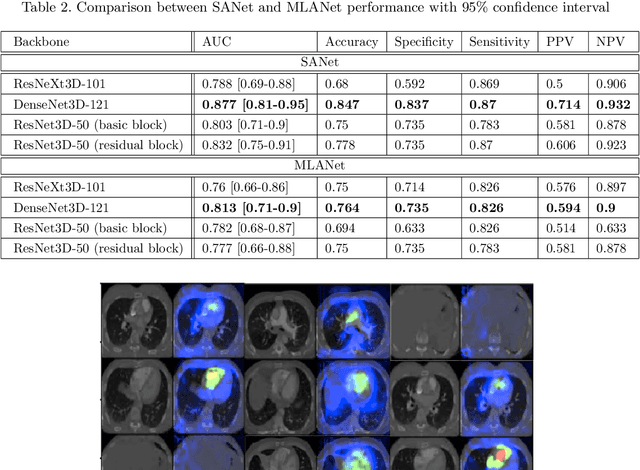
Abstract:Pulmonary embolus (PE) refers to obstruction of pulmonary arteries by blood clots. PE accounts for approximately 100,000 deaths per year in the United States alone. The clinical presentation of PE is often nonspecific, making the diagnosis challenging. Thus, rapid and accurate risk stratification is of paramount importance. High-risk PE is caused by right ventricular (RV) dysfunction from acute pressure overload, which in return can help identify which patients require more aggressive therapy. Reconstructed four-chamber views of the heart on chest CT can detect right ventricular enlargement. CT pulmonary angiography (CTPA) is the golden standard in the diagnostic workup of suspected PE. Therefore, it can link between diagnosis and risk stratification strategies. We developed a weakly supervised deep learning algorithm, with an emphasis on a novel attention mechanism, to automatically classify RV strain on CTPA. Our method is a 3D DenseNet model with integrated 3D residual attention blocks. We evaluated our model on a dataset of CTPAs of emergency department (ED) PE patients. This model achieved an area under the receiver operating characteristic curve (AUC) of 0.88 for classifying RV strain. The model showed a sensitivity of 87% and specificity of 83.7%. Our solution outperforms state-of-the-art 3D CNN networks. The proposed design allows for a fully automated network that can be trained easily in an end-to-end manner without requiring computationally intensive and time-consuming preprocessing or strenuous labeling of the data.We infer that unmarked CTPAs can be used for effective RV strain classification. This could be used as a second reader, alerting for high-risk PE patients. To the best of our knowledge, there are no previous deep learning-based studies that attempted to solve this problem.
Learning Rotation Invariant Features for Cryogenic Electron Microscopy Image Reconstruction
Jan 10, 2021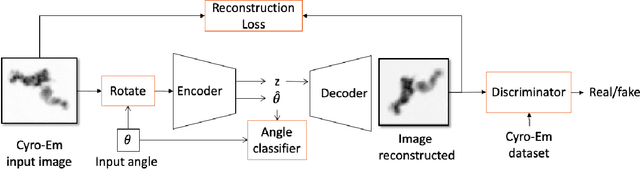
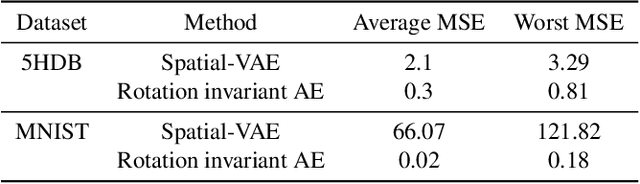
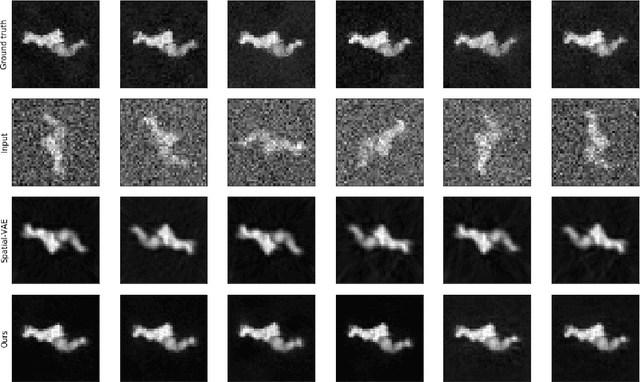
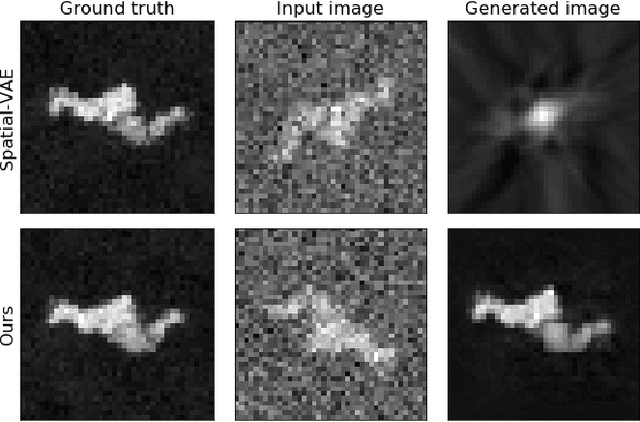
Abstract:Cryo-Electron Microscopy (Cryo-EM) is a Nobel prize-winning technology for determining the 3D structure of particles at near-atomic resolution. A fundamental step in the recovering of the 3D single-particle structure is to align its 2D projections; thus, the construction of a canonical representation with a fixed rotation angle is required. Most approaches use discrete clustering which fails to capture the continuous nature of image rotation, others suffer from low-quality image reconstruction. We propose a novel method that leverages the recent development in the generative adversarial networks. We introduce an encoder-decoder with a rotation angle classifier. In addition, we utilize a discriminator on the decoder output to minimize the reconstruction error. We demonstrate our approach with the Cryo-EM 5HDB and the rotated MNIST datasets showing substantial improvement over recent methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge