Eli Konen
X-ray2CTPA: Generating 3D CTPA scans from 2D X-ray conditioning
Jun 25, 2024Abstract:Chest X-rays or chest radiography (CXR), commonly used for medical diagnostics, typically enables limited imaging compared to computed tomography (CT) scans, which offer more detailed and accurate three-dimensional data, particularly contrast-enhanced scans like CT Pulmonary Angiography (CTPA). However, CT scans entail higher costs, greater radiation exposure, and are less accessible than CXRs. In this work we explore cross-modal translation from a 2D low contrast-resolution X-ray input to a 3D high contrast and spatial-resolution CTPA scan. Driven by recent advances in generative AI, we introduce a novel diffusion-based approach to this task. We evaluate the models performance using both quantitative metrics and qualitative feedback from radiologists, ensuring diagnostic relevance of the generated images. Furthermore, we employ the synthesized 3D images in a classification framework and show improved AUC in a PE categorization task, using the initial CXR input. The proposed method is generalizable and capable of performing additional cross-modality translations in medical imaging. It may pave the way for more accessible and cost-effective advanced diagnostic tools. The code for this project is available: https://github.com/NoaCahan/X-ray2CTPA .
Weakly Supervised Attention Model for RV StrainClassification from volumetric CTPA Scans
Jul 26, 2021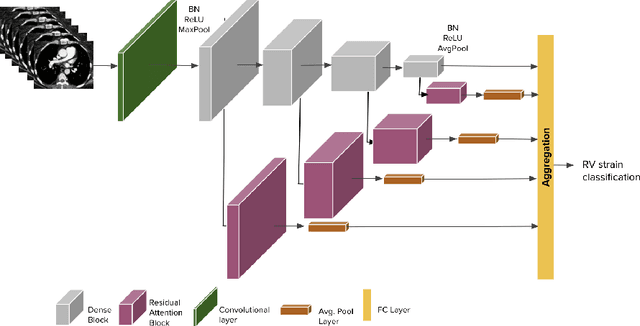
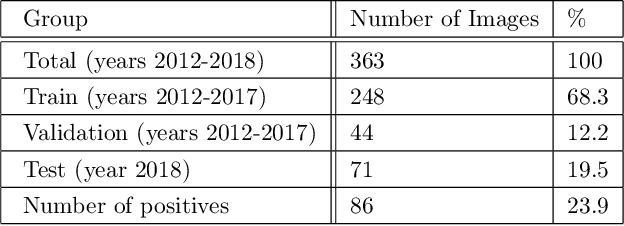
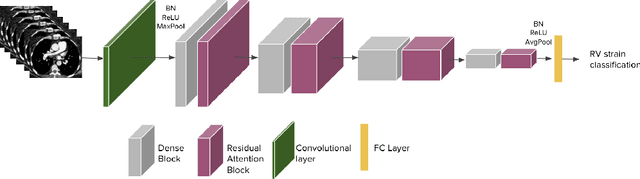
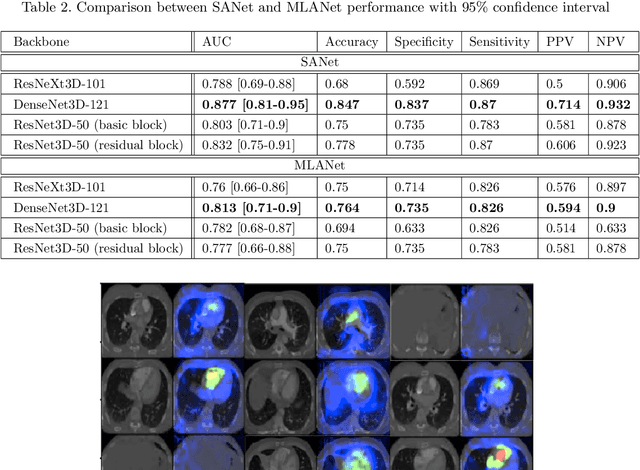
Abstract:Pulmonary embolus (PE) refers to obstruction of pulmonary arteries by blood clots. PE accounts for approximately 100,000 deaths per year in the United States alone. The clinical presentation of PE is often nonspecific, making the diagnosis challenging. Thus, rapid and accurate risk stratification is of paramount importance. High-risk PE is caused by right ventricular (RV) dysfunction from acute pressure overload, which in return can help identify which patients require more aggressive therapy. Reconstructed four-chamber views of the heart on chest CT can detect right ventricular enlargement. CT pulmonary angiography (CTPA) is the golden standard in the diagnostic workup of suspected PE. Therefore, it can link between diagnosis and risk stratification strategies. We developed a weakly supervised deep learning algorithm, with an emphasis on a novel attention mechanism, to automatically classify RV strain on CTPA. Our method is a 3D DenseNet model with integrated 3D residual attention blocks. We evaluated our model on a dataset of CTPAs of emergency department (ED) PE patients. This model achieved an area under the receiver operating characteristic curve (AUC) of 0.88 for classifying RV strain. The model showed a sensitivity of 87% and specificity of 83.7%. Our solution outperforms state-of-the-art 3D CNN networks. The proposed design allows for a fully automated network that can be trained easily in an end-to-end manner without requiring computationally intensive and time-consuming preprocessing or strenuous labeling of the data.We infer that unmarked CTPAs can be used for effective RV strain classification. This could be used as a second reader, alerting for high-risk PE patients. To the best of our knowledge, there are no previous deep learning-based studies that attempted to solve this problem.
Automatic Breast Lesion Classification by Joint Neural Analysis of Mammography and Ultrasound
Sep 23, 2020



Abstract:Mammography and ultrasound are extensively used by radiologists as complementary modalities to achieve better performance in breast cancer diagnosis. However, existing computer-aided diagnosis (CAD) systems for the breast are generally based on a single modality. In this work, we propose a deep-learning based method for classifying breast cancer lesions from their respective mammography and ultrasound images. We present various approaches and show a consistent improvement in performance when utilizing both modalities. The proposed approach is based on a GoogleNet architecture, fine-tuned for our data in two training steps. First, a distinct neural network is trained separately for each modality, generating high-level features. Then, the aggregated features originating from each modality are used to train a multimodal network to provide the final classification. In quantitative experiments, the proposed approach achieves an AUC of 0.94, outperforming state-of-the-art models trained over a single modality. Moreover, it performs similarly to an average radiologist, surpassing two out of four radiologists participating in a reader study. The promising results suggest that the proposed method may become a valuable decision support tool for breast radiologists.
Labeling of Multilingual Breast MRI Reports
Jul 06, 2020



Abstract:Medical reports are an essential medium in recording a patient's condition throughout a clinical trial. They contain valuable information that can be extracted to generate a large labeled dataset needed for the development of clinical tools. However, the majority of medical reports are stored in an unregularized format, and a trained human annotator (typically a doctor) must manually assess and label each case, resulting in an expensive and time consuming procedure. In this work, we present a framework for developing a multilingual breast MRI report classifier using a custom-built language representation called LAMBR. Our proposed method overcomes practical challenges faced in clinical settings, and we demonstrate improved performance in extracting labels from medical reports when compared with conventional approaches.
Knee Injury Detection using MRI with Efficiently-Layered Network (ELNet)
May 06, 2020



Abstract:Magnetic Resonance Imaging (MRI) is a widely-accepted imaging technique for knee injury analysis. Its advantage of capturing knee structure in three dimensions makes it the ideal tool for radiologists to locate potential tears in the knee. In order to better confront the ever growing workload of musculoskeletal (MSK) radiologists, automated tools for patients' triage are becoming a real need, reducing delays in the reading of pathological cases. In this work, we present the Efficiently-Layered Network (ELNet), a convolutional neural network (CNN) architecture optimized for the task of initial knee MRI diagnosis for triage. Unlike past approaches, we train ELNet from scratch instead of using a transfer-learning approach. The proposed method is validated quantitatively and qualitatively, and compares favorably against state-of-the-art MRNet while using a single imaging stack (axial or coronal) as input. Additionally, we demonstrate our model's capability to locate tears in the knee despite the absence of localization information during training. Lastly, the proposed model is extremely lightweight ($<$ 1MB) and therefore easy to train and deploy in real clinical settings.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019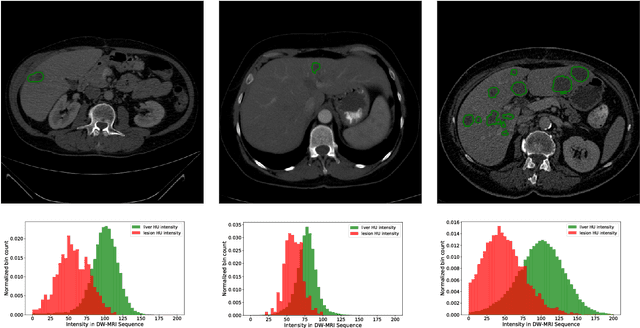
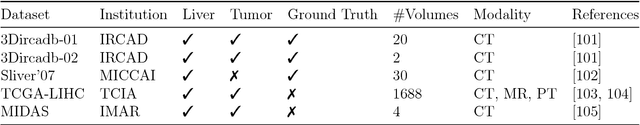
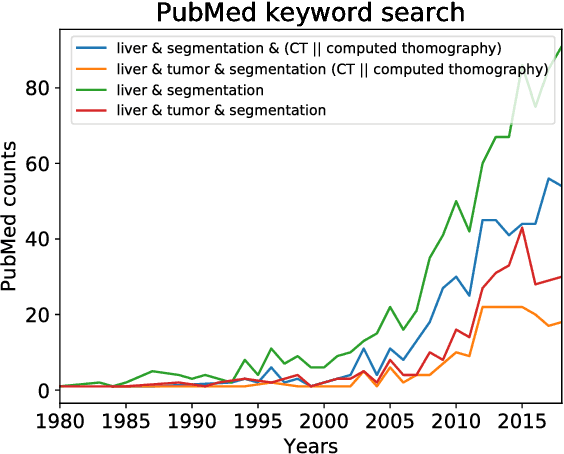
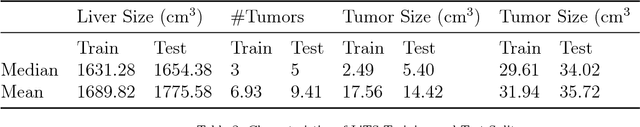
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Cross-Modality Synthesis from CT to PET using FCN and GAN Networks for Improved Automated Lesion Detection
Jul 23, 2018



Abstract:In this work we present a novel system for generation of virtual PET images using CT scans. We combine a fully convolutional network (FCN) with a conditional generative adversarial network (GAN) to generate simulated PET data from given input CT data. The synthesized PET can be used for false-positive reduction in lesion detection solutions. Clinically, such solutions may enable lesion detection and drug treatment evaluation in a CT-only environment, thus reducing the need for the more expensive and radioactive PET/CT scan. Our dataset includes 60 PET/CT scans from Sheba Medical center. We used 23 scans for training and 37 for testing. Different schemes to achieve the synthesized output were qualitatively compared. Quantitative evaluation was conducted using an existing lesion detection software, combining the synthesized PET as a false positive reduction layer for the detection of malignant lesions in the liver. Current results look promising showing a 28% reduction in the average false positive per case from 2.9 to 2.1. The suggested solution is comprehensive and can be expanded to additional body organs, and different modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge