Avi Ben-Cohen
It's All in the Head: Representation Knowledge Distillation through Classifier Sharing
Jan 18, 2022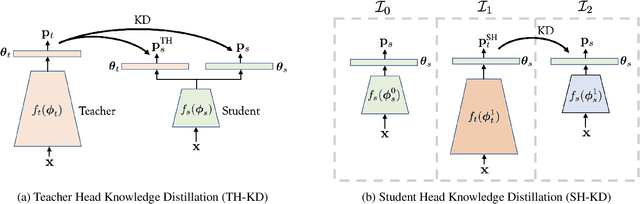
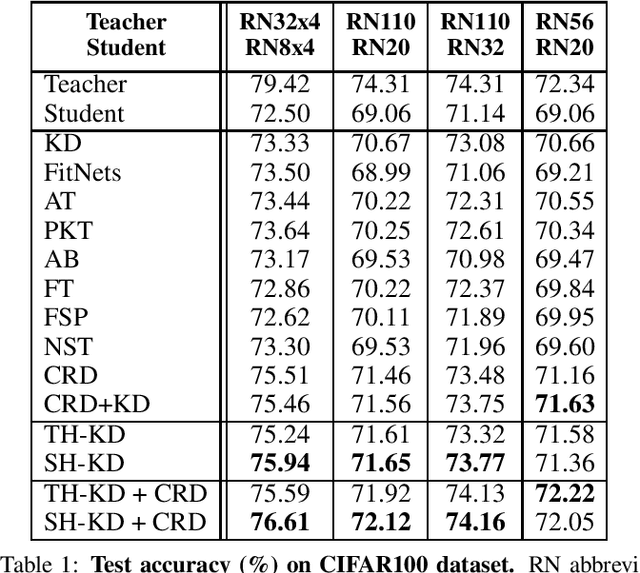
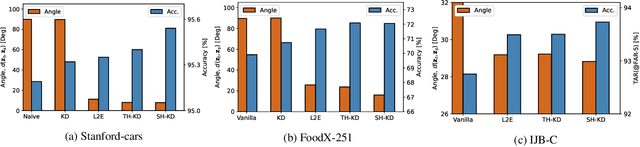
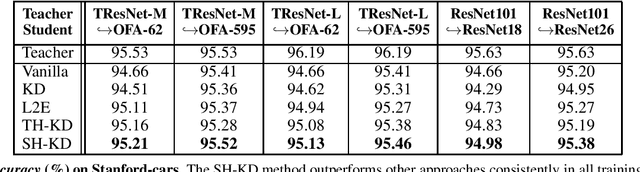
Abstract:Representation knowledge distillation aims at transferring rich information from one model to another. Current approaches for representation distillation mainly focus on the direct minimization of distance metrics between the models' embedding vectors. Such direct methods may be limited in transferring high-order dependencies embedded in the representation vectors, or in handling the capacity gap between the teacher and student models. In this paper, we introduce two approaches for enhancing representation distillation using classifier sharing between the teacher and student. Specifically, we first show that connecting the teacher's classifier to the student backbone and freezing its parameters is beneficial for the process of representation distillation, yielding consistent improvements. Then, we propose an alternative approach that asks to tailor the teacher model to a student with limited capacity. This approach competes with and in some cases surpasses the first method. Via extensive experiments and analysis, we show the effectiveness of the proposed methods on various datasets and tasks, including image classification, fine-grained classification, and face verification. For example, we achieve state-of-the-art performance for face verification on the IJB-C dataset for a MobileFaceNet model: TAR@(FAR=1e-5)=93.7\%. Code is available at https://github.com/Alibaba-MIIL/HeadSharingKD.
ML-Decoder: Scalable and Versatile Classification Head
Nov 25, 2021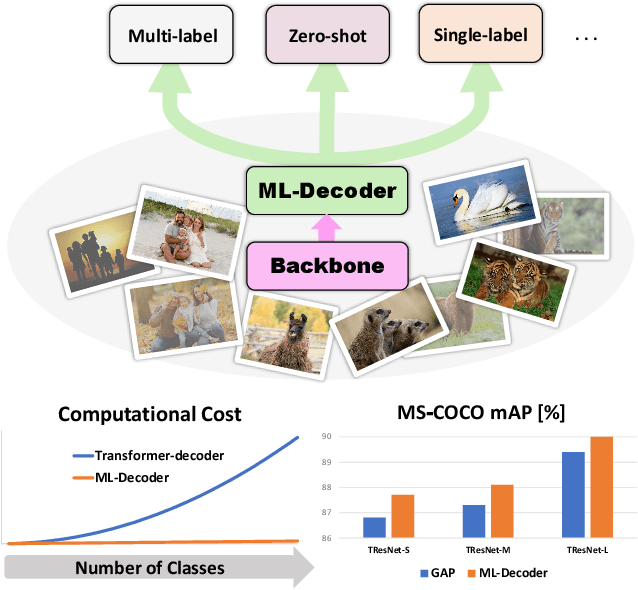
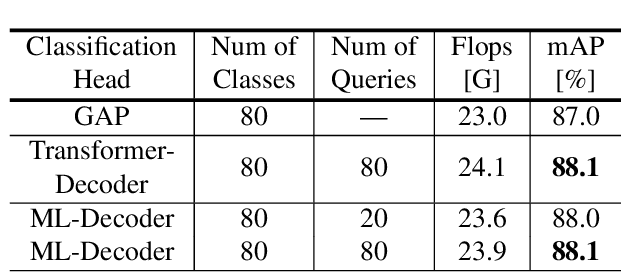
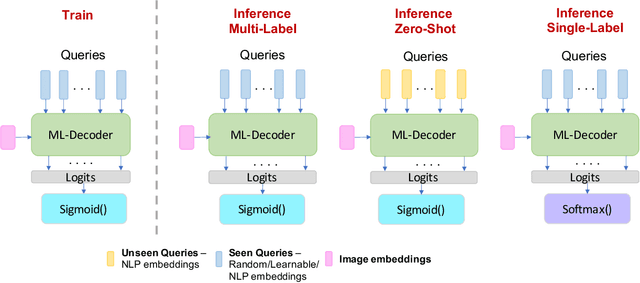
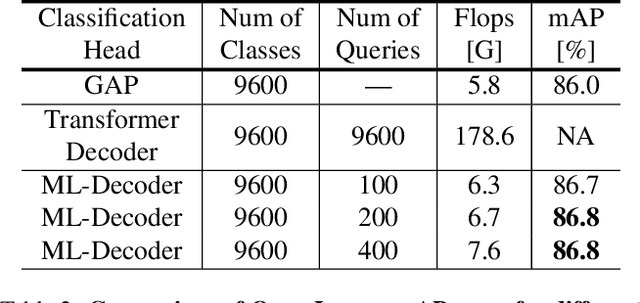
Abstract:In this paper, we introduce ML-Decoder, a new attention-based classification head. ML-Decoder predicts the existence of class labels via queries, and enables better utilization of spatial data compared to global average pooling. By redesigning the decoder architecture, and using a novel group-decoding scheme, ML-Decoder is highly efficient, and can scale well to thousands of classes. Compared to using a larger backbone, ML-Decoder consistently provides a better speed-accuracy trade-off. ML-Decoder is also versatile - it can be used as a drop-in replacement for various classification heads, and generalize to unseen classes when operated with word queries. Novel query augmentations further improve its generalization ability. Using ML-Decoder, we achieve state-of-the-art results on several classification tasks: on MS-COCO multi-label, we reach 91.4% mAP; on NUS-WIDE zero-shot, we reach 31.1% ZSL mAP; and on ImageNet single-label, we reach with vanilla ResNet50 backbone a new top score of 80.7%, without extra data or distillation. Public code is available at: https://github.com/Alibaba-MIIL/ML_Decoder
Multi-label Classification with Partial Annotations using Class-aware Selective Loss
Oct 21, 2021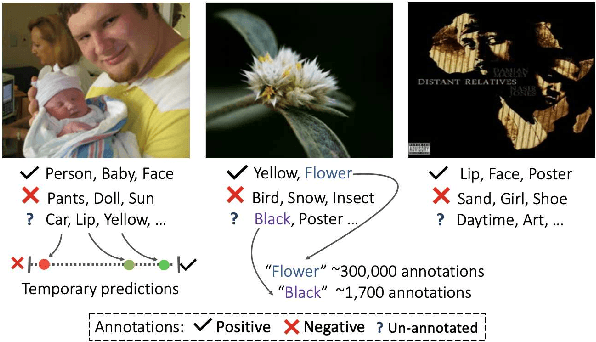
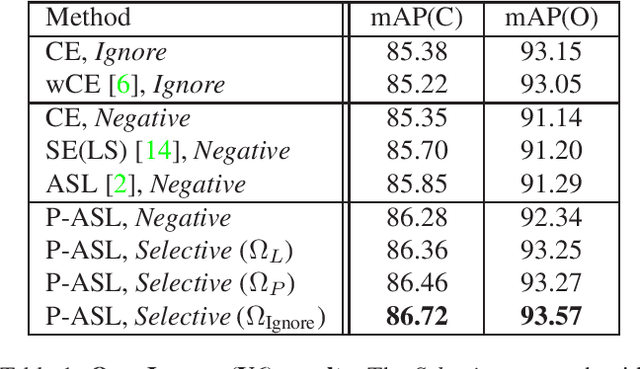
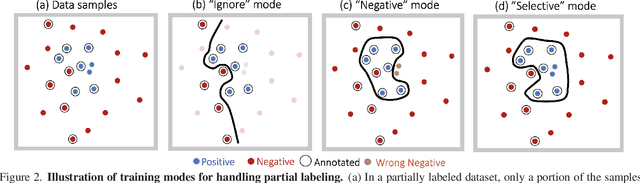
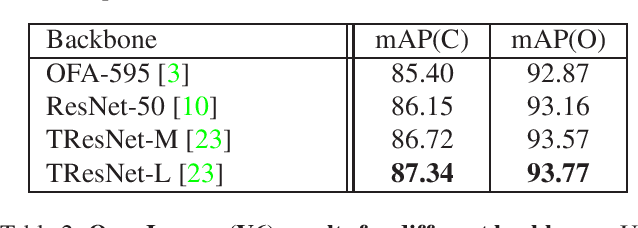
Abstract:Large-scale multi-label classification datasets are commonly, and perhaps inevitably, partially annotated. That is, only a small subset of labels are annotated per sample. Different methods for handling the missing labels induce different properties on the model and impact its accuracy. In this work, we analyze the partial labeling problem, then propose a solution based on two key ideas. First, un-annotated labels should be treated selectively according to two probability quantities: the class distribution in the overall dataset and the specific label likelihood for a given data sample. We propose to estimate the class distribution using a dedicated temporary model, and we show its improved efficiency over a naive estimation computed using the dataset's partial annotations. Second, during the training of the target model, we emphasize the contribution of annotated labels over originally un-annotated labels by using a dedicated asymmetric loss. With our novel approach, we achieve state-of-the-art results on OpenImages dataset (e.g. reaching 87.3 mAP on V6). In addition, experiments conducted on LVIS and simulated-COCO demonstrate the effectiveness of our approach. Code is available at https://github.com/Alibaba-MIIL/PartialLabelingCSL.
Semantic Diversity Learning for Zero-Shot Multi-label Classification
May 12, 2021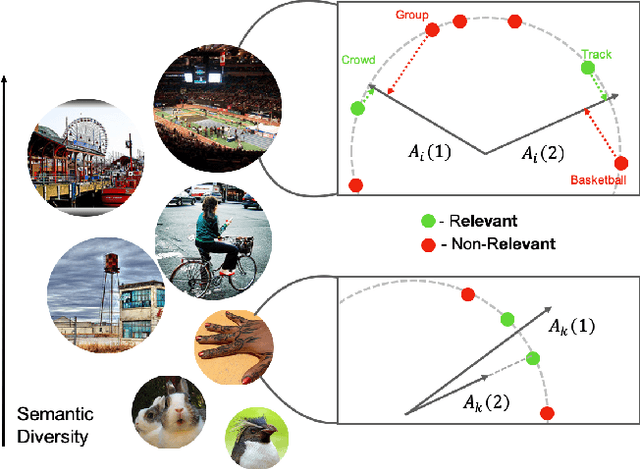
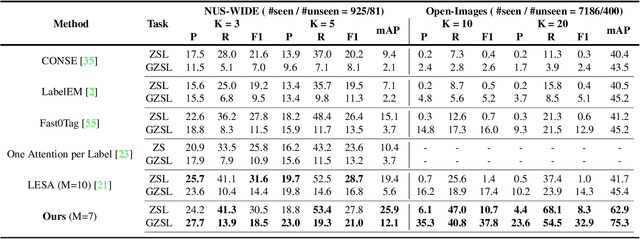
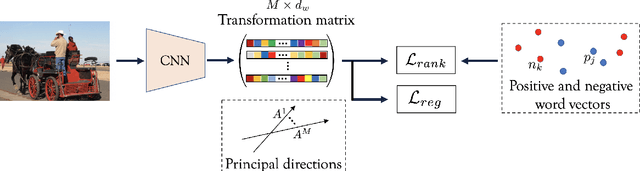
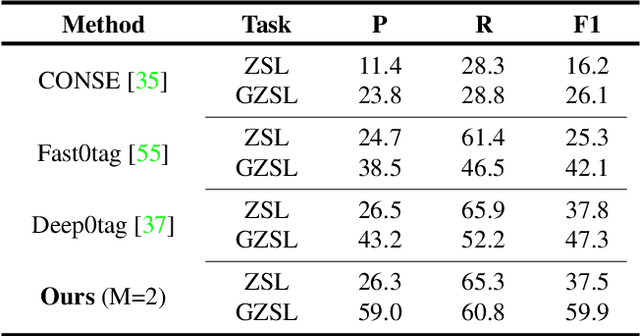
Abstract:Training a neural network model for recognizing multiple labels associated with an image, including identifying unseen labels, is challenging, especially for images that portray numerous semantically diverse labels. As challenging as this task is, it is an essential task to tackle since it represents many real-world cases, such as image retrieval of natural images. We argue that using a single embedding vector to represent an image, as commonly practiced, is not sufficient to rank both relevant seen and unseen labels accurately. This study introduces an end-to-end model training for multi-label zero-shot learning that supports semantic diversity of the images and labels. We propose to use an embedding matrix having principal embedding vectors trained using a tailored loss function. In addition, during training, we suggest up-weighting in the loss function image samples presenting higher semantic diversity to encourage the diversity of the embedding matrix. Extensive experiments show that our proposed method improves the zero-shot model's quality in tag-based image retrieval achieving SoTA results on several common datasets (NUS-Wide, COCO, Open Images).
Attention Network Robustification for Person ReID
Oct 29, 2019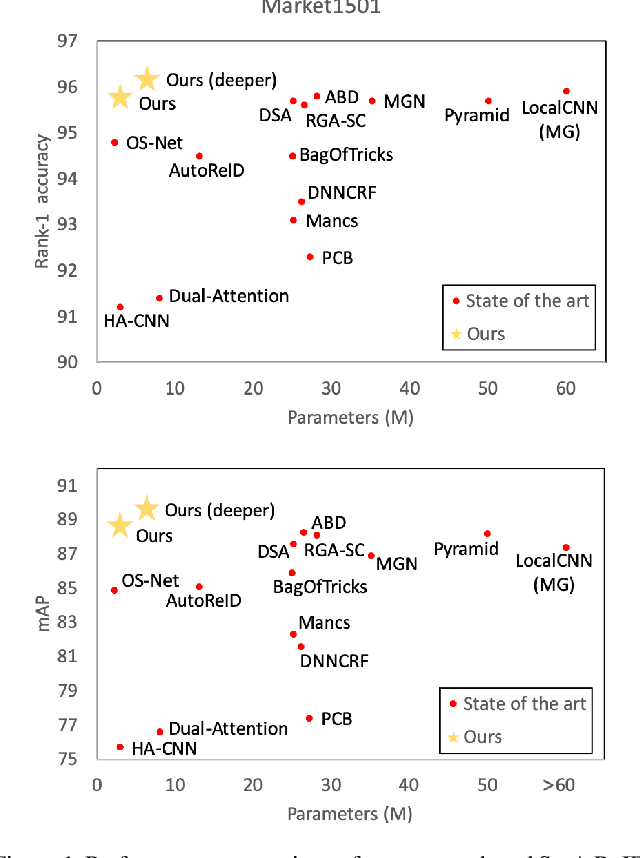
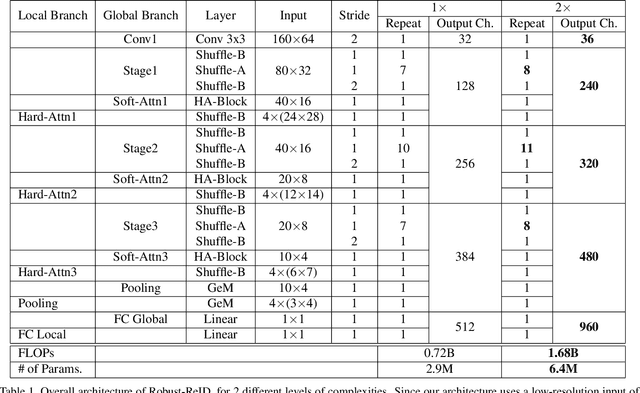
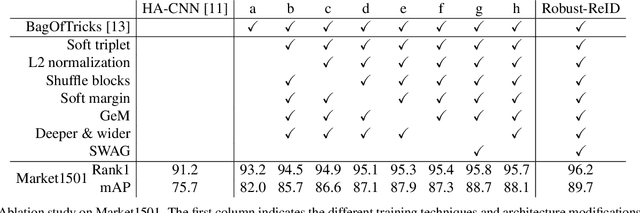
Abstract:The task of person re-identification (ReID) has attracted growing attention in recent years with improving performance but lack of focus on real-world applications. Most state of the art methods use large pre-trained models, e.g., ResNet50 (~25M parameters), as their backbone, which makes it tedious to explore different architecture modifications. In this study, we focus on small-sized randomly initialized models which enable us to easily introduce network and training modifications suitable for person ReID public datasets and real-world setups. We show the robustness of our network and training improvements by outperforming state of the art results in terms of rank-1 accuracy and mAP on Market1501 (96.2, 89.7) and DukeMTMC (89.8, 80.3) with only 6.4M parameters and without using re-ranking. Finally, we show the applicability of the proposed ReID network for multi-object tracking.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019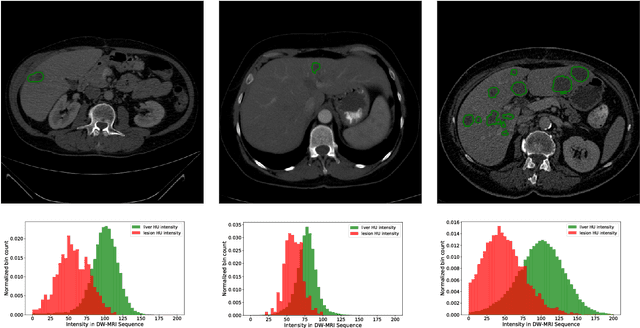
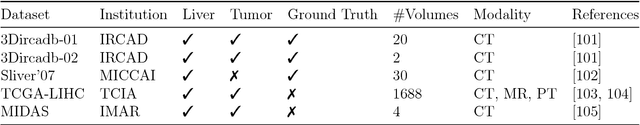
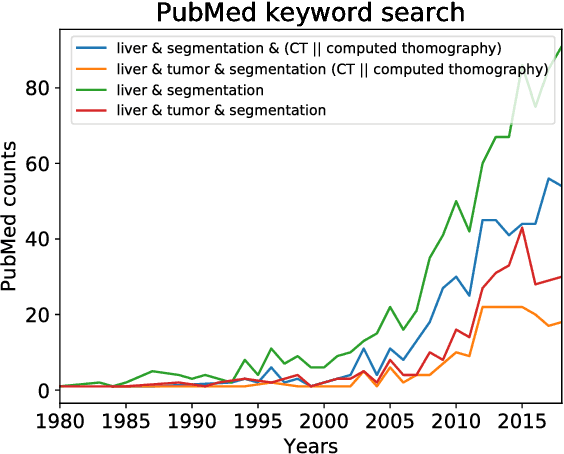
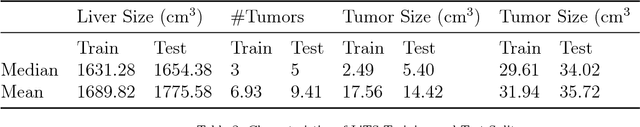
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Improving CNN Training using Disentanglement for Liver Lesion Classification in CT
Nov 01, 2018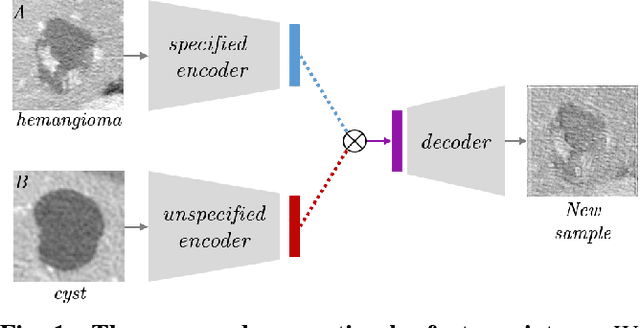
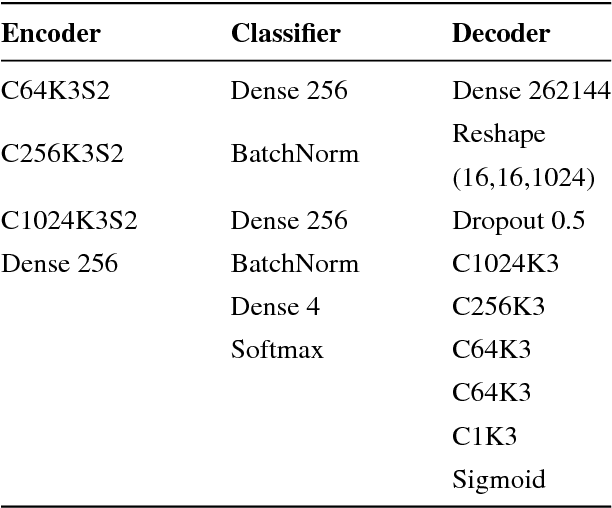
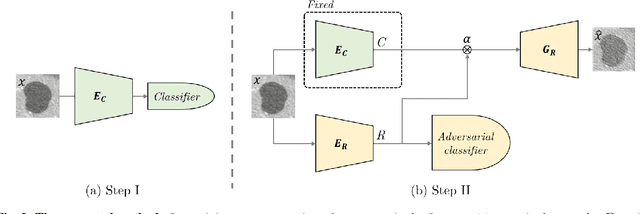
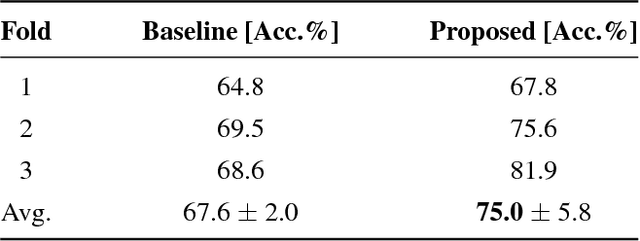
Abstract:Training data is the key component in designing algorithms for medical image analysis and in many cases it is the main bottleneck in achieving good results. Recent progress in image generation has enabled the training of neural network based solutions using synthetic data. A key factor in the generation of new samples is controlling the important appearance features and potentially being able to generate a new sample of a specific class with different variants. In this work we suggest the synthesis of new data by mixing the class specified and unspecified representation of different factors in the training data. Our experiments on liver lesion classification in CT show an average improvement of 7.4% in accuracy over the baseline training scheme.
Cross-Modality Synthesis from CT to PET using FCN and GAN Networks for Improved Automated Lesion Detection
Jul 23, 2018



Abstract:In this work we present a novel system for generation of virtual PET images using CT scans. We combine a fully convolutional network (FCN) with a conditional generative adversarial network (GAN) to generate simulated PET data from given input CT data. The synthesized PET can be used for false-positive reduction in lesion detection solutions. Clinically, such solutions may enable lesion detection and drug treatment evaluation in a CT-only environment, thus reducing the need for the more expensive and radioactive PET/CT scan. Our dataset includes 60 PET/CT scans from Sheba Medical center. We used 23 scans for training and 37 for testing. Different schemes to achieve the synthesized output were qualitatively compared. Quantitative evaluation was conducted using an existing lesion detection software, combining the synthesized PET as a false positive reduction layer for the detection of malignant lesions in the liver. Current results look promising showing a 28% reduction in the average false positive per case from 2.9 to 2.1. The suggested solution is comprehensive and can be expanded to additional body organs, and different modalities.
Anatomical Data Augmentation For CNN based Pixel-wise Classification
Jan 07, 2018



Abstract:In this work we propose a method for anatomical data augmentation that is based on using slices of computed tomography (CT) examinations that are adjacent to labeled slices as another resource of labeled data for training the network. The extended labeled data is used to train a U-net network for a pixel-wise classification into different hepatic lesions and normal liver tissues. Our dataset contains CT examinations from 140 patients with 333 CT images annotated by an expert radiologist. We tested our approach and compared it to the conventional training process. Results indicate superiority of our method. Using the anatomical data augmentation we achieved an improvement of 3% in the success rate, 5% in the classification accuracy, and 4% in Dice.
Virtual PET Images from CT Data Using Deep Convolutional Networks: Initial Results
Jul 30, 2017



Abstract:In this work we present a novel system for PET estimation using CT scans. We explore the use of fully convolutional networks (FCN) and conditional generative adversarial networks (GAN) to export PET data from CT data. Our dataset includes 25 pairs of PET and CT scans where 17 were used for training and 8 for testing. The system was tested for detection of malignant tumors in the liver region. Initial results look promising showing high detection performance with a TPR of 92.3% and FPR of 0.25 per case. Future work entails expansion of the current system to the entire body using a much larger dataset. Such a system can be used for tumor detection and drug treatment evaluation in a CT-only environment instead of the expansive and radioactive PET-CT scan.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge