Jürgen Hesser
A baseline for machine-learning-based hepatocellular carcinoma diagnosis using multi-modal clinical data
Jan 20, 2025Abstract:The objective of this paper is to provide a baseline for performing multi-modal data classification on a novel open multimodal dataset of hepatocellular carcinoma (HCC), which includes both image data (contrast-enhanced CT and MRI images) and tabular data (the clinical laboratory test data as well as case report forms). TNM staging is the classification task. Features from the vectorized preprocessed tabular data and radiomics features from contrast-enhanced CT and MRI images are collected. Feature selection is performed based on mutual information. An XGBoost classifier predicts the TNM staging and it shows a prediction accuracy of $0.89 \pm 0.05$ and an AUC of $0.93 \pm 0.03$. The classifier shows that this high level of prediction accuracy can only be obtained by combining image and clinical laboratory data and therefore is a good example case where multi-model classification is mandatory to achieve accurate results.
SynBench: A Synthetic Benchmark for Non-rigid 3D Point Cloud Registration
Sep 22, 2024



Abstract:Non-rigid point cloud registration is a crucial task in computer vision. Evaluating a non-rigid point cloud registration method requires a dataset with challenges such as large deformation levels, noise, outliers, and incompleteness. Despite the existence of several datasets for deformable point cloud registration, the absence of a comprehensive benchmark with all challenges makes it difficult to achieve fair evaluations among different methods. This paper introduces SynBench, a new non-rigid point cloud registration dataset created using SimTool, a toolset for soft body simulation in Flex and Unreal Engine. SynBench provides the ground truth of corresponding points between two point sets and encompasses key registration challenges, including varying levels of deformation, noise, outliers, and incompleteness. To the best of the authors' knowledge, compared to existing datasets, SynBench possesses three particular characteristics: (1) it is the first benchmark that provides various challenges for non-rigid point cloud registration, (2) SynBench encompasses challenges of varying difficulty levels, and (3) it includes ground truth corresponding points both before and after deformation. The authors believe that SynBench enables future non-rigid point cloud registration methods to present a fair comparison of their achievements. SynBench is publicly available at: https://doi.org/10.11588/data/R9IKCF.
Weighted Monte Carlo augmented spherical Fourier-Bessel convolutional layers for 3D abdominal organ segmentation
Mar 09, 2024



Abstract:Filter-decomposition-based group equivariant convolutional neural networks show promising stability and data efficiency for 3D image feature extraction. However, the existing filter-decomposition-based 3D group equivariant neural networks rely on parameter-sharing designs and are mostly limited to rotation transformation groups, where the chosen spherical harmonic filter bases consider only angular orthogonality. These limitations hamper its application to deep neural network architectures for medical image segmentation. To address these issues, this paper describes a non-parameter-sharing affine group equivariant neural network for 3D medical image segmentation based on an adaptive aggregation of Monte Carlo augmented spherical Fourier Bessel filter bases. The efficiency and flexibility of the adopted non-parameter-sharing strategy enable for the first time an efficient implementation of 3D affine group equivariant convolutional neural networks for volumetric data. The introduced spherical Bessel Fourier filter basis combines both angular and radial orthogonality for better feature extraction. The 3D image segmentation experiments on two abdominal medical image sets, BTCV and the NIH Pancreas datasets, show that the proposed methods excel the state-of-the-art 3D neural networks with high training stability and data efficiency. The code will be available at https://github.com/ZhaoWenzhao/WMCSFB.
Robust-DefReg: A Robust Deformable Point Cloud Registration Method based on Graph Convolutional Neural Networks
Jun 07, 2023



Abstract:Point cloud registration is a fundamental problem in computer vision that aims to estimate the transformation between corresponding sets of points. Non-rigid registration, in particular, involves addressing challenges including various levels of deformation, noise, outliers, and data incompleteness. This paper introduces Robust-DefReg, a robust non-rigid point cloud registration method based on graph convolutional networks (GCNNs). Robust-DefReg is a coarse-to-fine registration approach within an end-to-end pipeline, leveraging the advantages of both coarse and fine methods. The method learns global features to find correspondences between source and target point clouds, to enable appropriate initial alignment, and subsequently fine registration. The simultaneous achievement of high accuracy and robustness across all challenges is reported less frequently in existing studies, making it a key objective of the Robust-DefReg method. The proposed method achieves high accuracy in large deformations while maintaining computational efficiency. This method possesses three primary attributes: high accuracy, robustness to different challenges, and computational efficiency. The experimental results show that the proposed Robust-DefReg holds significant potential as a foundational architecture for future investigations in non-rigid point cloud registration. The source code of Robust-DefReg is available.
Adaptive aggregation of Monte Carlo augmented decomposed filters for efficient group-equivariant convolutional neural network
May 17, 2023



Abstract:Filter-decomposition-based group-equivariant convolutional neural networks (G-CNN) have been demonstrated to increase CNN's data efficiency and contribute to better interpretability and controllability of CNN models. However, so far filter-decomposition-based affine G-CNN methods rely on parameter sharing for achieving high parameter efficiency and suffer from a heavy computational burden. They also use a limited number of transformations and in particular ignore the shear transform in the application. In this paper, we address these problems by emphasizing the importance of the diversity of transformations. We propose a flexible and efficient strategy based on weighted filter-wise Monte Carlo sampling. In addition, we introduce shear equivariant CNN to address the highly sparse representations of natural images. We demonstrate that the proposed methods are intrinsically an efficient generalization of traditional CNNs, and we explain the advantage of bottleneck architectures used in the existing state-of-the-art CNN models such as ResNet, ResNext, and ConvNeXt from the group-equivariant perspective. Experiments on image classification and image denoising tasks show that with a set of suitable filter basis, our methods achieve superior performance to standard CNN with high data efficiency. The code will be available at https://github.com/ZhaoWenzhao/MCG_CNN.
Boosting Liver and Lesion Segmentation from CT Scans By Mask Mining
Aug 14, 2019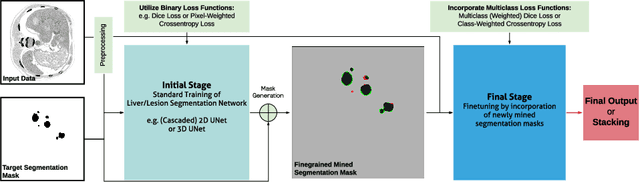


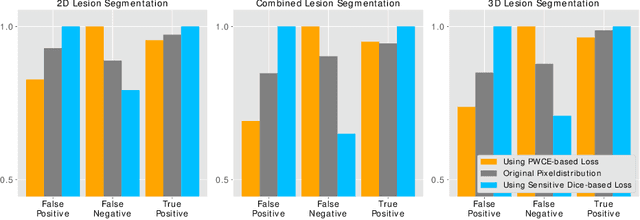
Abstract:In this paper we propose a novel procedure to improve liver and liver lesion segmentation from CT scans for U-Net based models. Our method is an extension to standard segmentation pipelines allowing for more fine-grained control over the network output by focusing on higher target recall or reduction of noisy false-positive predictions, thereby also boosting overall segmentation performance. To achieve this, we include segmentation errors after a primary learning step into a new learning process appended to the main training setup, allowing the model to find features which explain away previous errors. We evaluate this on distinct architectures including cascaded two- and three-dimensional as well as combined learning setups for multitask segmentation. Liver and lesion segmentation data is provided by the Liver Tumor Segmentationchallenge (LiTS), with an increase in dice score of up to 3 points.
Liver Lesion Segmentation with slice-wise 2D Tiramisu and Tversky loss function
May 09, 2019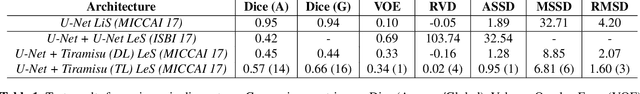

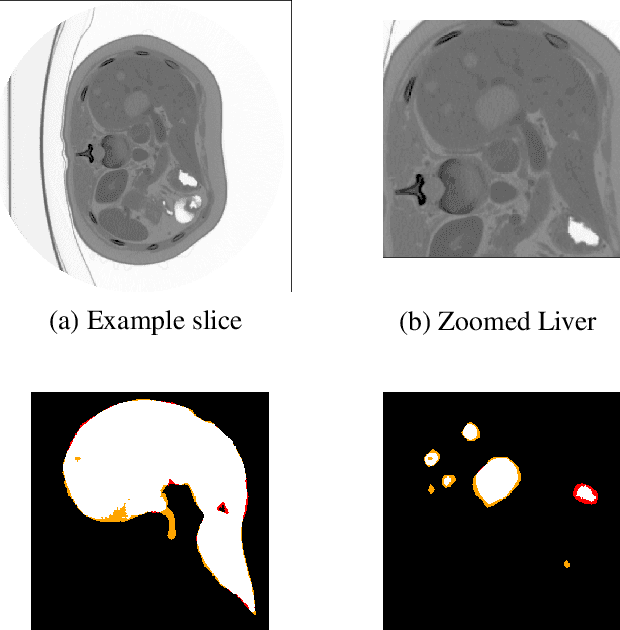
Abstract:At present, lesion segmentation is still performed manually (or semi-automatically) by medical experts. To facilitate this process, we contribute a fully-automatic lesion segmentation pipeline. This work proposes a method as a part of the LiTS (Liver Tumor Segmentation Challenge) competition for ISBI 17 and MICCAI 17 comparing methods for automatics egmentation of liver lesions in CT scans. By utilizing cascaded, densely connected 2D U-Nets and a Tversky-coefficient based loss function, our framework achieves very good shape extractions with high detection sensitivity, with competitive scores at time of publication. In addition, adjusting hyperparameters in our Tversky-loss allows to tune the network towards higher sensitivity or robustness.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019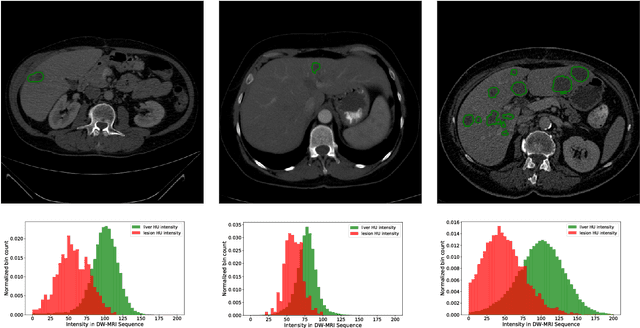
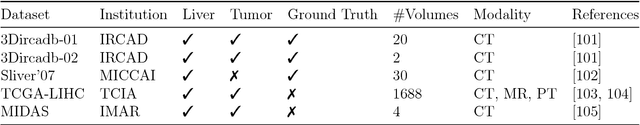
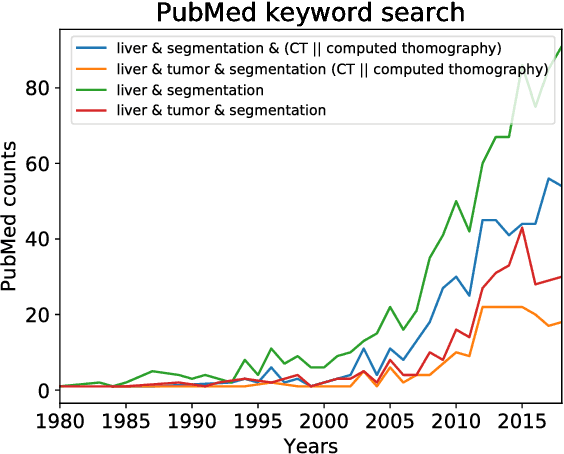
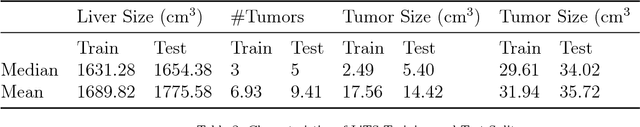
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Automated Multiscale 3D Feature Learning for Vessels Segmentation in Thorax CT Images
Jan 06, 2019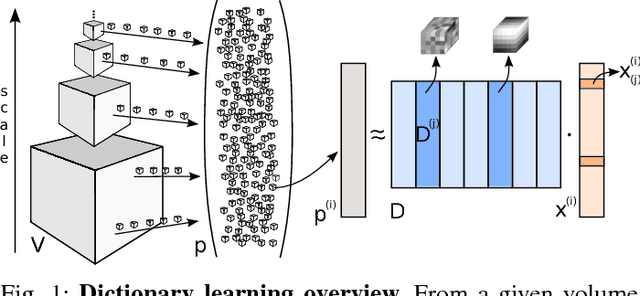
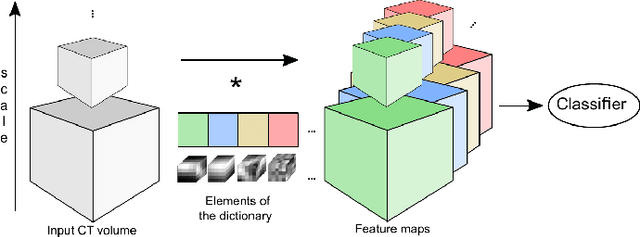


Abstract:We address the vessel segmentation problem by building upon the multiscale feature learning method of Kiros et al., which achieves the current top score in the VESSEL12 MICCAI challenge. Following their idea of feature learning instead of hand-crafted filters, we have extended the method to learn 3D features. The features are learned in an unsupervised manner in a multi-scale scheme using dictionary learning via least angle regression. The 3D feature kernels are further convolved with the input volumes in order to create feature maps. Those maps are used to train a supervised classifier with the annotated voxels. In order to process the 3D data with a large number of filters a parallel implementation has been developed. The algorithm has been applied on the example scans and annotations provided by the VESSEL12 challenge. We have compared our setup with Kiros et al. by running their implementation. Our current results show an improvement in accuracy over the slice wise method from 96.66$\pm$1.10% to 97.24$\pm$0.90%.
Fully Convolutional Deep Network Architectures for Automatic Short Glass Fiber Semantic Segmentation from CT scans
Jan 04, 2019



Abstract:We present the first attempt to perform short glass fiber semantic segmentation from X-ray computed tomography volumetric datasets at medium (3.9 {\mu}m isotropic) and low (8.3 {\mu}m isotropic) resolution using deep learning architectures. We performed experiments on both synthetic and real CT scans and evaluated deep fully convolutional architectures with both 2D and 3D kernels. Our artificial neural networks outperform existing methods at both medium and low resolution scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge