Tomasz Konopczyński
Boosting Liver and Lesion Segmentation from CT Scans By Mask Mining
Aug 14, 2019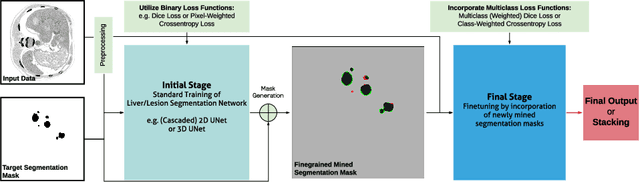


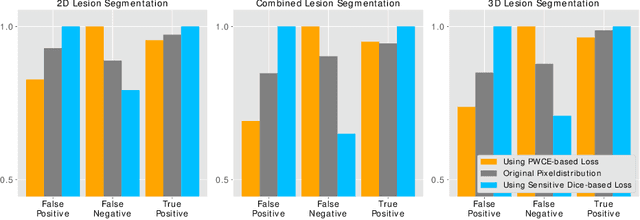
Abstract:In this paper we propose a novel procedure to improve liver and liver lesion segmentation from CT scans for U-Net based models. Our method is an extension to standard segmentation pipelines allowing for more fine-grained control over the network output by focusing on higher target recall or reduction of noisy false-positive predictions, thereby also boosting overall segmentation performance. To achieve this, we include segmentation errors after a primary learning step into a new learning process appended to the main training setup, allowing the model to find features which explain away previous errors. We evaluate this on distinct architectures including cascaded two- and three-dimensional as well as combined learning setups for multitask segmentation. Liver and lesion segmentation data is provided by the Liver Tumor Segmentationchallenge (LiTS), with an increase in dice score of up to 3 points.
Liver Lesion Segmentation with slice-wise 2D Tiramisu and Tversky loss function
May 09, 2019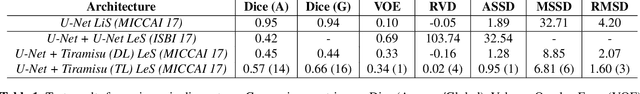

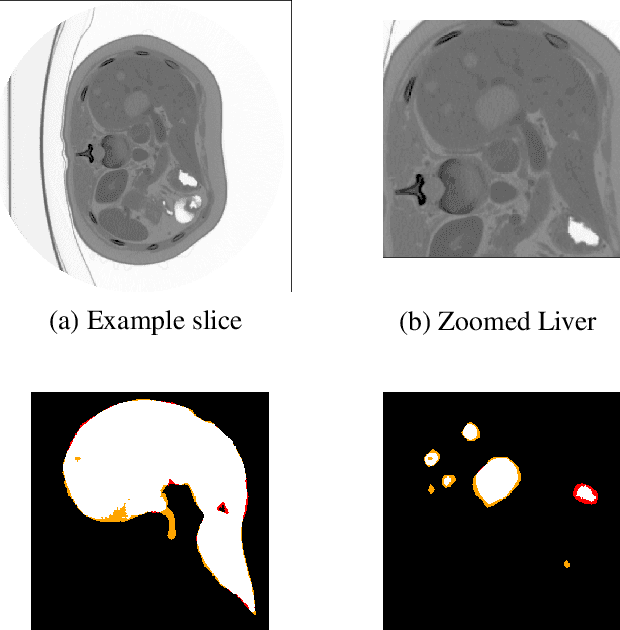
Abstract:At present, lesion segmentation is still performed manually (or semi-automatically) by medical experts. To facilitate this process, we contribute a fully-automatic lesion segmentation pipeline. This work proposes a method as a part of the LiTS (Liver Tumor Segmentation Challenge) competition for ISBI 17 and MICCAI 17 comparing methods for automatics egmentation of liver lesions in CT scans. By utilizing cascaded, densely connected 2D U-Nets and a Tversky-coefficient based loss function, our framework achieves very good shape extractions with high detection sensitivity, with competitive scores at time of publication. In addition, adjusting hyperparameters in our Tversky-loss allows to tune the network towards higher sensitivity or robustness.
Automated Multiscale 3D Feature Learning for Vessels Segmentation in Thorax CT Images
Jan 06, 2019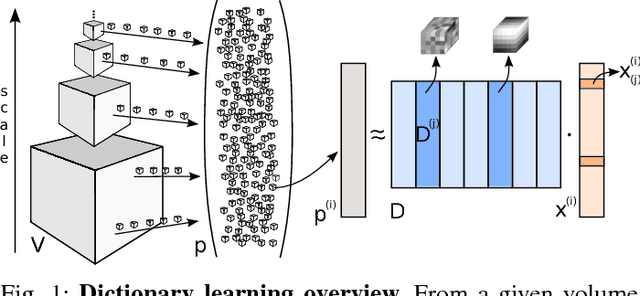
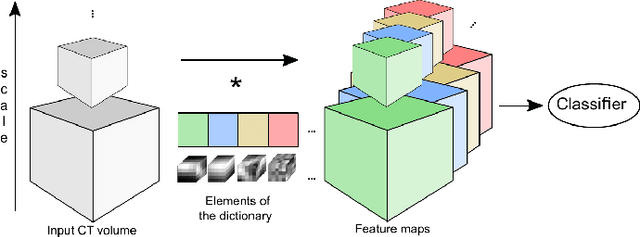


Abstract:We address the vessel segmentation problem by building upon the multiscale feature learning method of Kiros et al., which achieves the current top score in the VESSEL12 MICCAI challenge. Following their idea of feature learning instead of hand-crafted filters, we have extended the method to learn 3D features. The features are learned in an unsupervised manner in a multi-scale scheme using dictionary learning via least angle regression. The 3D feature kernels are further convolved with the input volumes in order to create feature maps. Those maps are used to train a supervised classifier with the annotated voxels. In order to process the 3D data with a large number of filters a parallel implementation has been developed. The algorithm has been applied on the example scans and annotations provided by the VESSEL12 challenge. We have compared our setup with Kiros et al. by running their implementation. Our current results show an improvement in accuracy over the slice wise method from 96.66$\pm$1.10% to 97.24$\pm$0.90%.
Fully Convolutional Deep Network Architectures for Automatic Short Glass Fiber Semantic Segmentation from CT scans
Jan 04, 2019



Abstract:We present the first attempt to perform short glass fiber semantic segmentation from X-ray computed tomography volumetric datasets at medium (3.9 {\mu}m isotropic) and low (8.3 {\mu}m isotropic) resolution using deep learning architectures. We performed experiments on both synthetic and real CT scans and evaluated deep fully convolutional architectures with both 2D and 3D kernels. Our artificial neural networks outperform existing methods at both medium and low resolution scans.
Reference Setup for Quantitative Comparison of Segmentation Techniques for Short Glass Fiber CT Data
Jan 04, 2019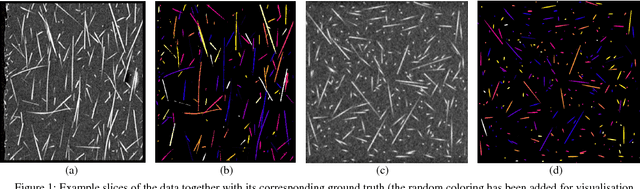
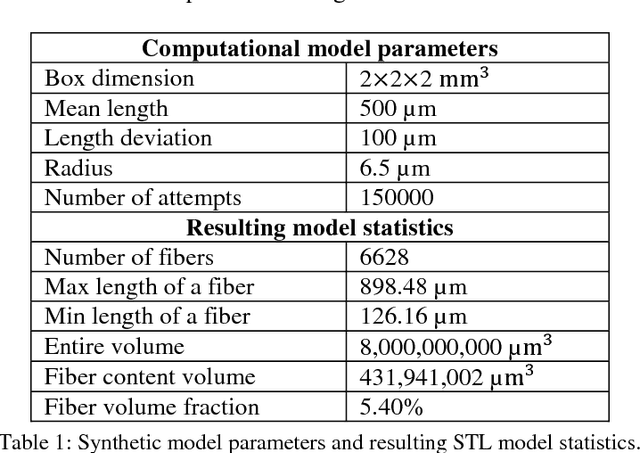
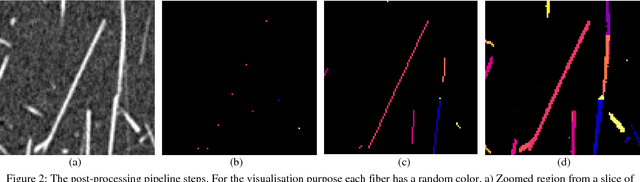
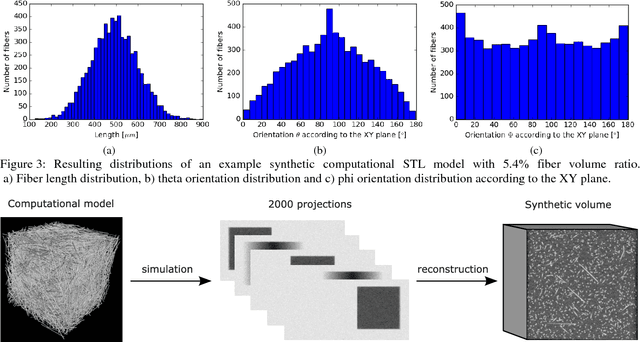
Abstract:Comparing different algorithms for segmenting glass fibers in industrial computed tomography (CT) scans is difficult due to the absence of a standard reference dataset. In this work, we introduce a set of annotated scans of short-fiber reinforced polymers (SFRP) as well as synthetically created CT volume data together with the evaluation metrics. We suggest both the metrics and this data set as a reference for studying the performance of different algorithms. The real scans were acquired by a Nikon MCT225 X-ray CT system. The simulated scans were created by the use of an in-house computational model and third-party commercial software. For both types of data, corresponding ground truth annotations have been prepared, including hand annotations for the real scans and STL models for the synthetic scans. Additionally, a Hessian-based Frangi vesselness filter for fiber segmentation has been implemented and open-sourced to serve as a reference for comparisons.
Instance Segmentation of Fibers from Low Resolution CT Scans via 3D Deep Embedding Learning
Jan 04, 2019



Abstract:We propose a novel approach for automatic extraction (instance segmentation) of fibers from low resolution 3D X-ray computed tomography scans of short glass fiber reinforced polymers. We have designed a 3D instance segmentation architecture built upon a deep fully convolutional network for semantic segmentation with an extra output for embedding learning. We show that the embedding learning is capable of learning a mapping of voxels to an embedded space in which a standard clustering algorithm can be used to distinguish between different instances of an object in a volume. In addition, we discuss a merging post-processing method which makes it possible to process volumes of any size. The proposed 3D instance segmentation network together with our merging algorithm is the first known to authors knowledge procedure that produces results good enough, that they can be used for further analysis of low resolution fiber composites CT scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge