Gili Weiss-Dicker
Unsupervised particle sorting for cryo-EM using probabilistic PCA
Oct 23, 2022Abstract:Single-particle cryo-electron microscopy (cryo-EM) is a leading technology to resolve the structure of molecules. Early in the process, the user detects potential particle images in the raw data. Typically, there are many false detections as a result of high levels of noise and contamination. Currently, removing the false detections requires human intervention to sort the hundred thousands of images. We propose a statistically-established unsupervised algorithm to remove non-particle images. We model the particle images as a union of low-dimensional subspaces, assuming non-particle images are arbitrarily scattered in the high-dimensional space. The algorithm is based on an extension of the probabilistic PCA framework to robustly learn a non-linear model of union of subspaces. This provides a flexible model for cryo-EM data, and allows to automatically remove images that correspond to pure noise and contamination. Numerical experiments corroborate the effectiveness of the sorting algorithm.
Learning Rotation Invariant Features for Cryogenic Electron Microscopy Image Reconstruction
Jan 10, 2021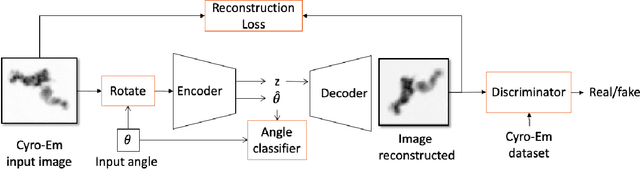
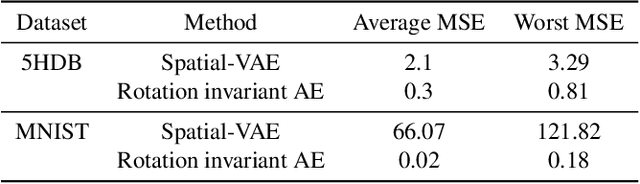
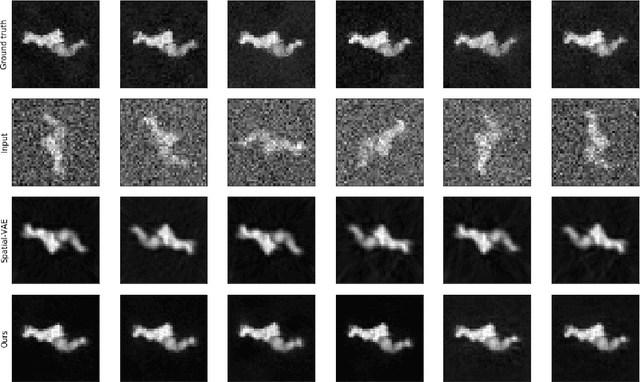
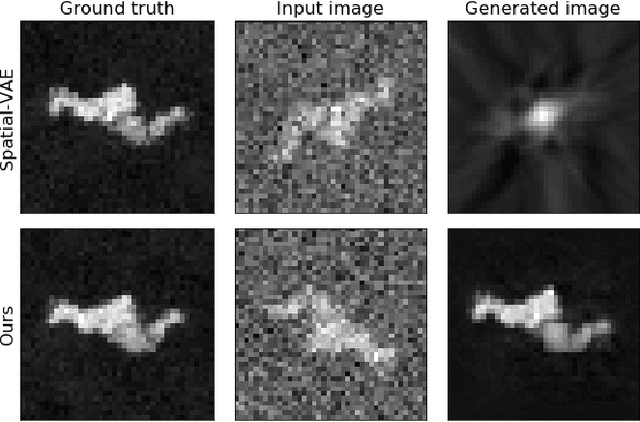
Abstract:Cryo-Electron Microscopy (Cryo-EM) is a Nobel prize-winning technology for determining the 3D structure of particles at near-atomic resolution. A fundamental step in the recovering of the 3D single-particle structure is to align its 2D projections; thus, the construction of a canonical representation with a fixed rotation angle is required. Most approaches use discrete clustering which fails to capture the continuous nature of image rotation, others suffer from low-quality image reconstruction. We propose a novel method that leverages the recent development in the generative adversarial networks. We introduce an encoder-decoder with a rotation angle classifier. In addition, we utilize a discriminator on the decoder output to minimize the reconstruction error. We demonstrate our approach with the Cryo-EM 5HDB and the rotated MNIST datasets showing substantial improvement over recent methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge