Amitay Eldar
Unsupervised particle sorting for cryo-EM using probabilistic PCA
Oct 23, 2022Abstract:Single-particle cryo-electron microscopy (cryo-EM) is a leading technology to resolve the structure of molecules. Early in the process, the user detects potential particle images in the raw data. Typically, there are many false detections as a result of high levels of noise and contamination. Currently, removing the false detections requires human intervention to sort the hundred thousands of images. We propose a statistically-established unsupervised algorithm to remove non-particle images. We model the particle images as a union of low-dimensional subspaces, assuming non-particle images are arbitrarily scattered in the high-dimensional space. The algorithm is based on an extension of the probabilistic PCA framework to robustly learn a non-linear model of union of subspaces. This provides a flexible model for cryo-EM data, and allows to automatically remove images that correspond to pure noise and contamination. Numerical experiments corroborate the effectiveness of the sorting algorithm.
ASOCEM: Automatic Segmentation Of Contaminations in cryo-EM
Jan 18, 2022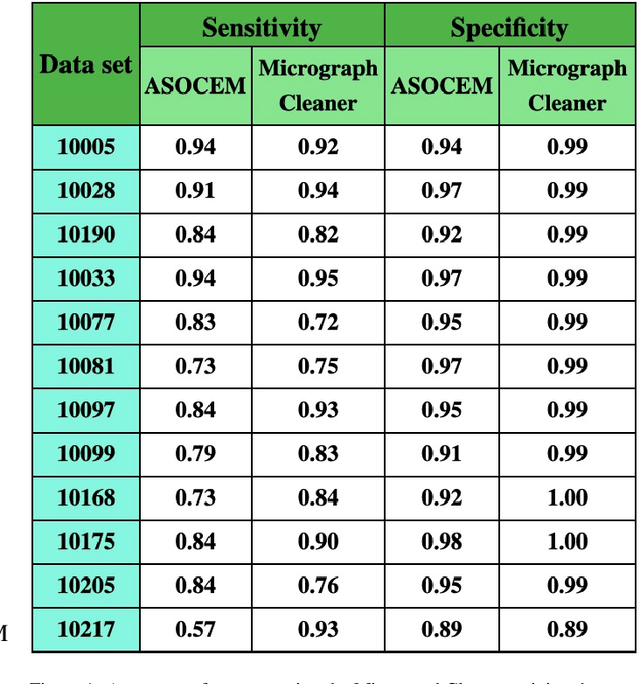
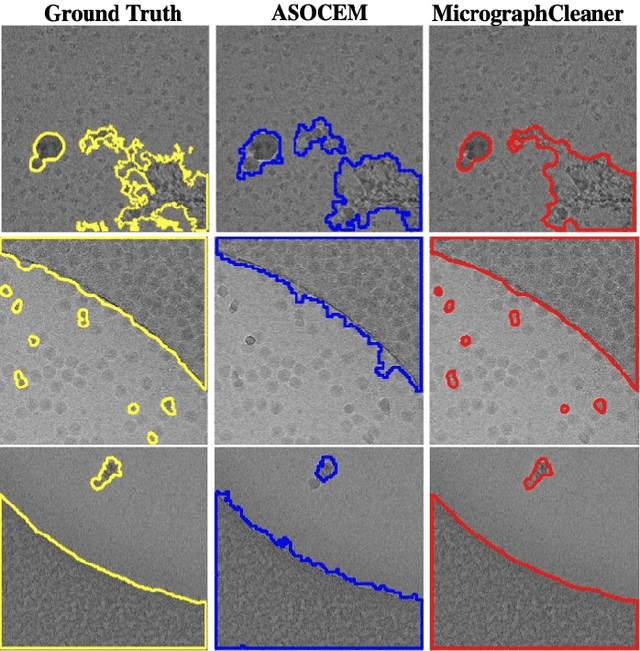
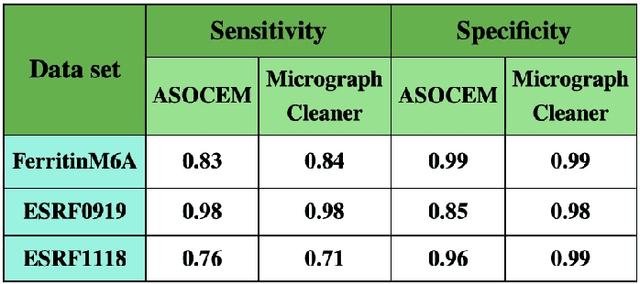

Abstract:Particle picking is currently a critical step in the cryo-electron microscopy single particle reconstruction pipeline. Contaminations in the acquired micrographs severely degrade the performance of particle pickers, resulting is many ``non-particles'' in the collected stack of particles. In this paper, we present ASOCEM (Automatic Segmentation Of Contaminations in cryo-EM), an automatic method to detect and segment contaminations, which requires as an input only the approximated particle size. In particular, it does not require any parameter tuning nor manual intervention. Our method is based on the observation that the statistical distribution of contaminated regions is different from that of the rest of the micrograph. This nonrestrictive assumption allows to automatically detect various types of contaminations, from the carbon edges of the supporting grid to high contrast blobs of different sizes. We demonstrate the efficiency of our algorithm using various experimental data sets containing various types of contaminations. ASOCEM is integrated as part of the KLT picker \cite{ELDAR2020107473} and is available at \url{https://github.com/ShkolniskyLab/kltpicker2}.
KLT Picker: Particle Picking Using Data-Driven Optimal Templates
Dec 12, 2019



Abstract:Particle picking is currently a critical step in the cryo-EM single particle reconstruction pipeline. Despite extensive work on this problem, for many data sets it is still challenging, especially for low SNR micrographs. We present the KLT (Karhunen Loeve Transform) picker, which is fully automatic and requires as an input only the approximated particle size. In particular, it does not require any manual picking. Our method is designed especially to handle low SNR micrographs. It is based on learning a set of optimal templates through the use of multi-variate statistical analysis via the Karhunen Loeve Transform. We evaluate the KLT picker on publicly available data sets and present high-quality results with minimal manual effort.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge