Maayan Frid-Adar
Automated triage of COVID-19 from various lung abnormalities using chest CT features
Oct 24, 2020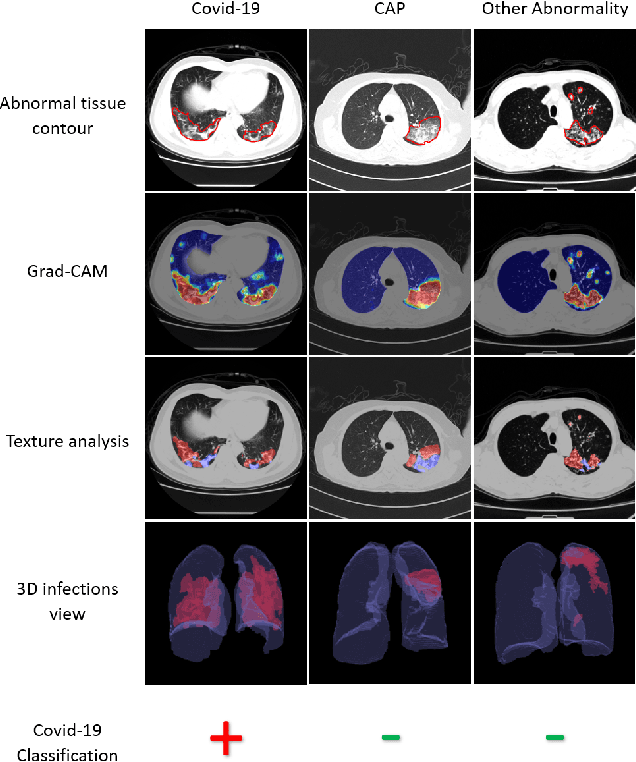

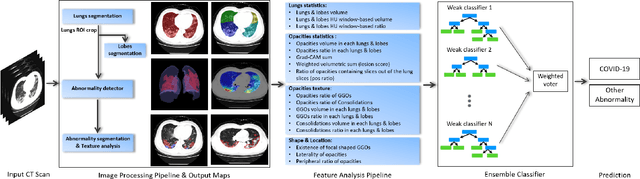
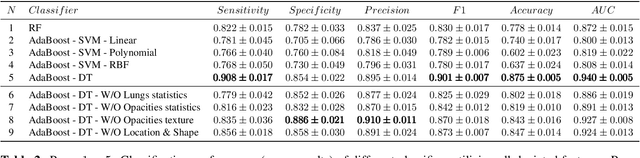
Abstract:The outbreak of COVID-19 has lead to a global effort to decelerate the pandemic spread. For this purpose chest computed-tomography (CT) based screening and diagnosis of COVID-19 suspected patients is utilized, either as a support or replacement to reverse transcription-polymerase chain reaction (RT-PCR) test. In this paper, we propose a fully automated AI based system that takes as input chest CT scans and triages COVID-19 cases. More specifically, we produce multiple descriptive features, including lung and infections statistics, texture, shape and location, to train a machine learning based classifier that distinguishes between COVID-19 and other lung abnormalities (including community acquired pneumonia). We evaluated our system on a dataset of 2191 CT cases and demonstrated a robust solution with 90.8% sensitivity at 85.4% specificity with 94.0% ROC-AUC. In addition, we present an elaborated feature analysis and ablation study to explore the importance of each feature.
COVID-19 in CXR: from Detection and Severity Scoring to Patient Disease Monitoring
Aug 04, 2020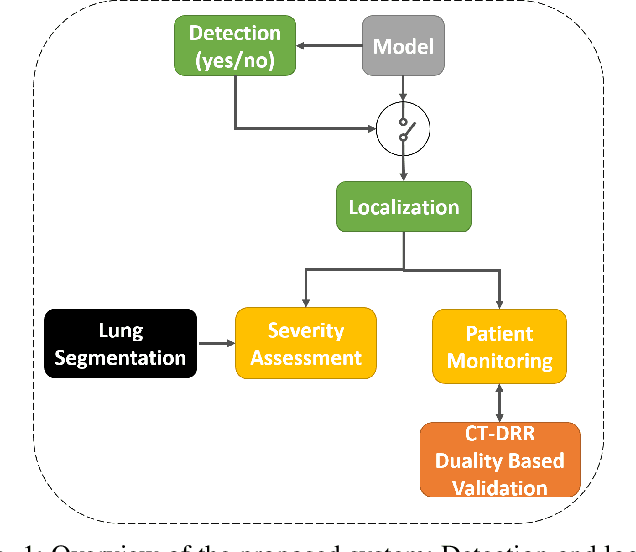
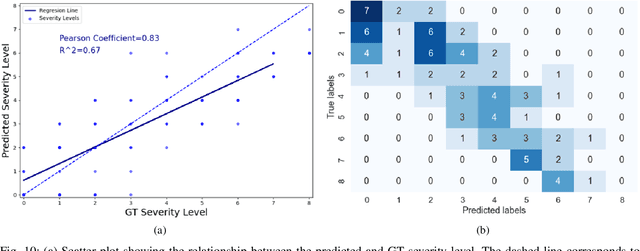
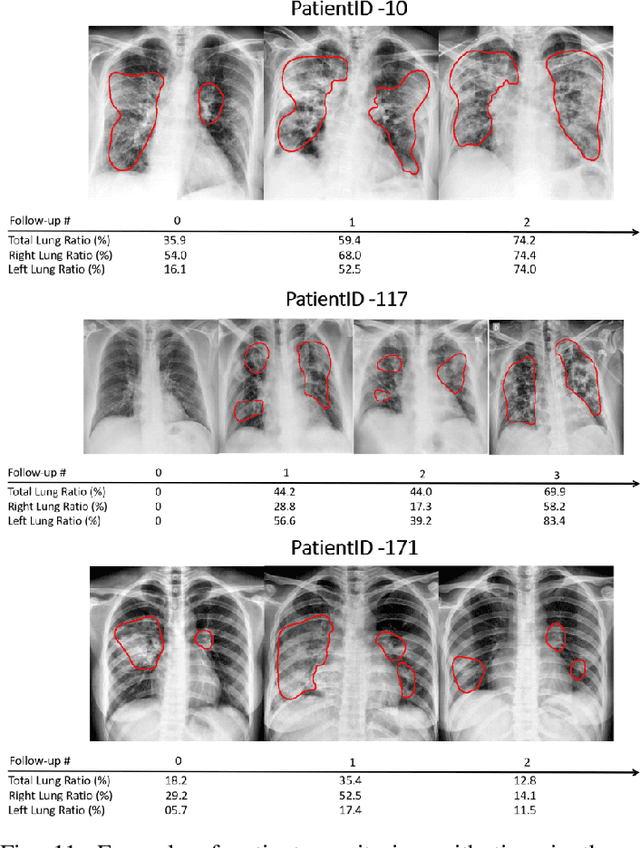
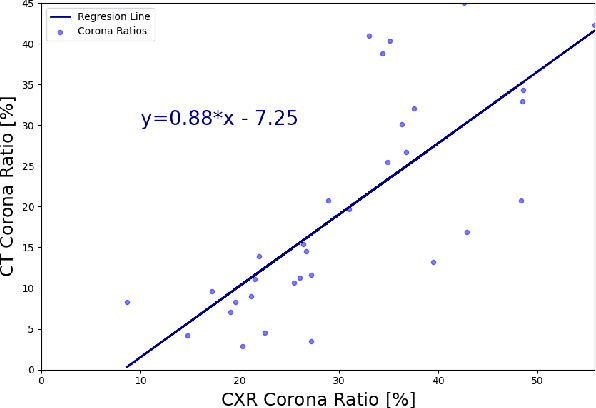
Abstract:In this work, we estimate the severity of pneumonia in COVID-19 patients and conduct a longitudinal study of disease progression. To achieve this goal, we developed a deep learning model for simultaneous detection and segmentation of pneumonia in chest Xray (CXR) images and generalized to COVID-19 pneumonia. The segmentations were utilized to calculate a "Pneumonia Ratio" which indicates the disease severity. The measurement of disease severity enables to build a disease extent profile over time for hospitalized patients. To validate the model relevance to the patient monitoring task, we developed a validation strategy which involves a synthesis of Digital Reconstructed Radiographs (DRRs - synthetic Xray) from serial CT scans; we then compared the disease progression profiles that were generated from the DRRs to those that were generated from CT volumes.
Coronavirus Detection and Analysis on Chest CT with Deep Learning
Apr 06, 2020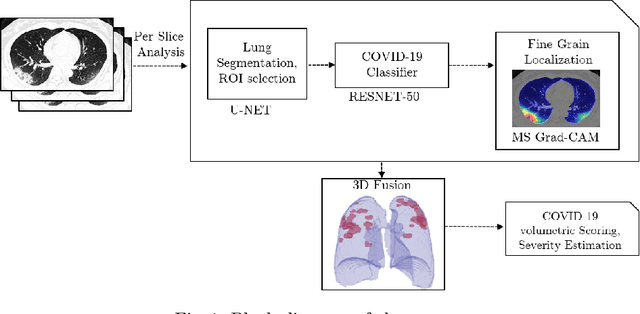

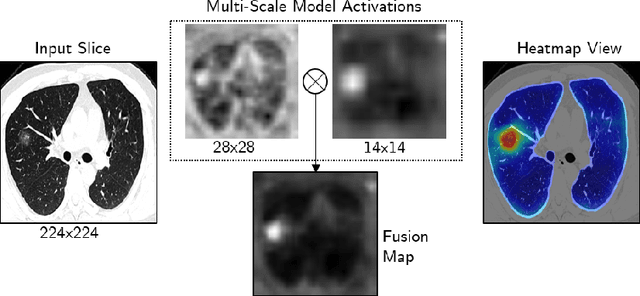
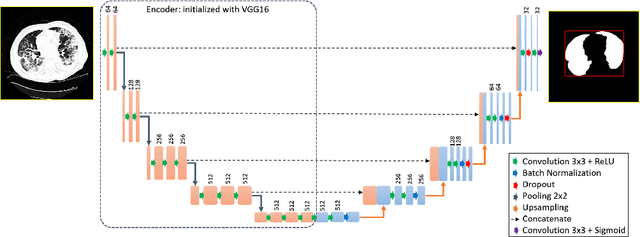
Abstract:The outbreak of the novel coronavirus, officially declared a global pandemic, has a severe impact on our daily lives. As of this writing there are approximately 197,188 confirmed cases of which 80,881 are in "Mainland China" with 7,949 deaths, a mortality rate of 3.4%. In order to support radiologists in this overwhelming challenge, we develop a deep learning based algorithm that can detect, localize and quantify severity of COVID-19 manifestation from chest CT scans. The algorithm is comprised of a pipeline of image processing algorithms which includes lung segmentation, 2D slice classification and fine grain localization. In order to further understand the manifestations of the disease, we perform unsupervised clustering of abnormal slices. We present our results on a dataset comprised of 110 confirmed COVID-19 patients from Zhejiang province, China.
Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis
Mar 24, 2020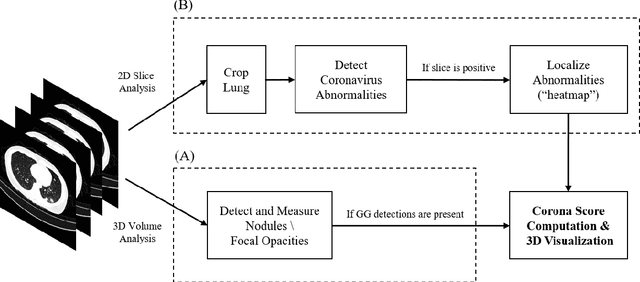
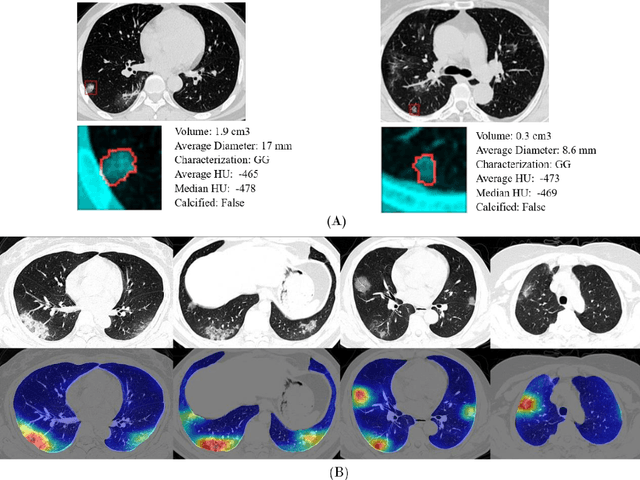
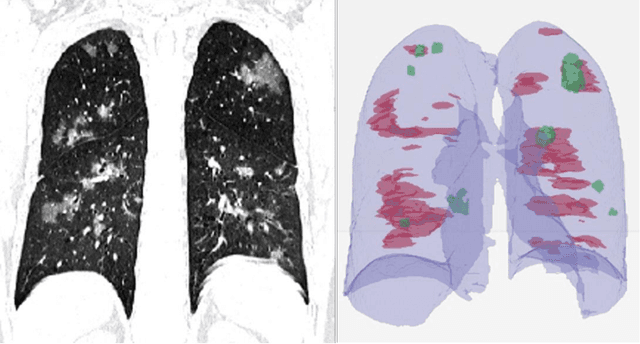
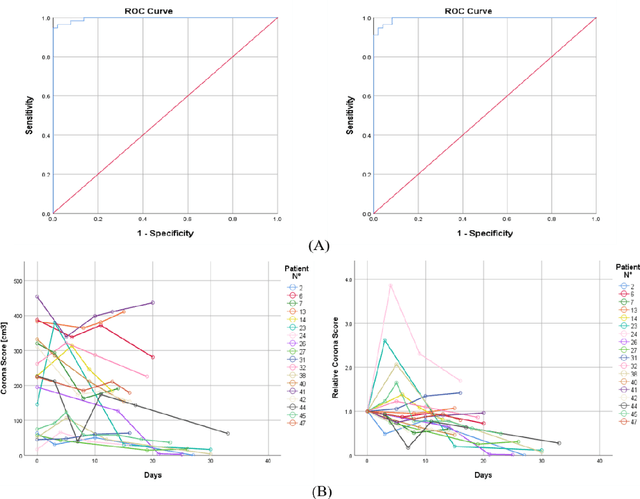
Abstract:Purpose: Develop AI-based automated CT image analysis tools for detection, quantification, and tracking of Coronavirus; demonstrate they can differentiate coronavirus patients from non-patients. Materials and Methods: Multiple international datasets, including from Chinese disease-infected areas were included. We present a system that utilizes robust 2D and 3D deep learning models, modifying and adapting existing AI models and combining them with clinical understanding. We conducted multiple retrospective experiments to analyze the performance of the system in the detection of suspected COVID-19 thoracic CT features and to evaluate evolution of the disease in each patient over time using a 3D volume review, generating a Corona score. The study includes a testing set of 157 international patients (China and U.S). Results: Classification results for Coronavirus vs Non-coronavirus cases per thoracic CT studies were 0.996 AUC (95%CI: 0.989-1.00) ; on datasets of Chinese control and infected patients. Possible working point: 98.2% sensitivity, 92.2% specificity. For time analysis of Coronavirus patients, the system output enables quantitative measurements for smaller opacities (volume, diameter) and visualization of the larger opacities in a slice-based heat map or a 3D volume display. Our suggested Corona score measures the progression of disease over time. Conclusion: This initial study, which is currently being expanded to a larger population, demonstrated that rapidly developed AI-based image analysis can achieve high accuracy in detection of Coronavirus as well as quantification and tracking of disease burden.
Endotracheal Tube Detection and Segmentation in Chest Radiographs using Synthetic Data
Aug 20, 2019



Abstract:Chest radiographs are frequently used to verify the correct intubation of patients in the emergency room. Fast and accurate identification and localization of the endotracheal (ET) tube is critical for the patient. In this study we propose a novel automated deep learning scheme for accurate detection and segmentation of the ET tubes. Development of automatic systems using deep learning networks for classification and segmentation require large annotated data which is not always available. Here we present an approach for synthesizing ET tubes in real X-ray images. We suggest a method for training the network, first with synthetic data and then with real X-ray images in a fine-tuning phase, which allows the network to train on thousands of cases without annotating any data. The proposed method was tested on 477 real chest radiographs from a public dataset and reached AUC of 0.99 in classifying the presence vs. absence of the ET tube, along with outputting high quality ET tube segmentation maps.
GAN-based Synthetic Medical Image Augmentation for increased CNN Performance in Liver Lesion Classification
Mar 03, 2018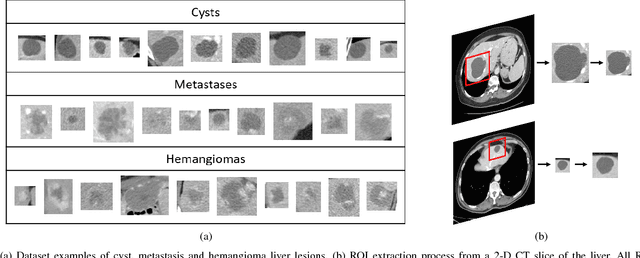
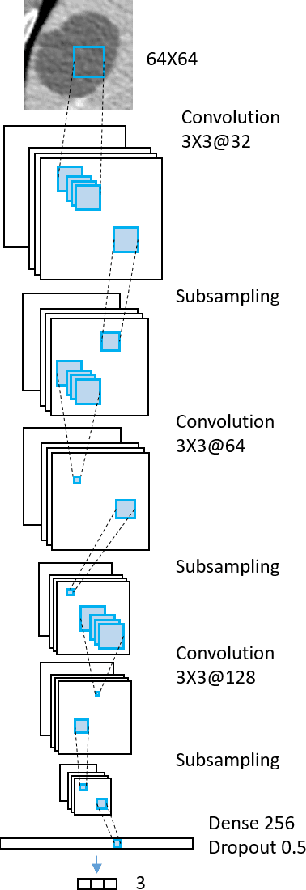
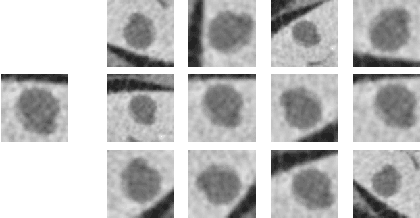
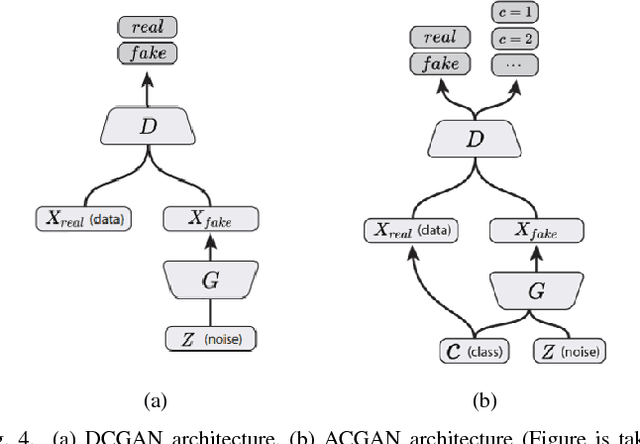
Abstract:Deep learning methods, and in particular convolutional neural networks (CNNs), have led to an enormous breakthrough in a wide range of computer vision tasks, primarily by using large-scale annotated datasets. However, obtaining such datasets in the medical domain remains a challenge. In this paper, we present methods for generating synthetic medical images using recently presented deep learning Generative Adversarial Networks (GANs). Furthermore, we show that generated medical images can be used for synthetic data augmentation, and improve the performance of CNN for medical image classification. Our novel method is demonstrated on a limited dataset of computed tomography (CT) images of 182 liver lesions (53 cysts, 64 metastases and 65 hemangiomas). We first exploit GAN architectures for synthesizing high quality liver lesion ROIs. Then we present a novel scheme for liver lesion classification using CNN. Finally, we train the CNN using classic data augmentation and our synthetic data augmentation and compare performance. In addition, we explore the quality of our synthesized examples using visualization and expert assessment. The classification performance using only classic data augmentation yielded 78.6% sensitivity and 88.4% specificity. By adding the synthetic data augmentation the results increased to 85.7% sensitivity and 92.4% specificity. We believe that this approach to synthetic data augmentation can generalize to other medical classification applications and thus support radiologists' efforts to improve diagnosis.
Synthetic Data Augmentation using GAN for Improved Liver Lesion Classification
Jan 08, 2018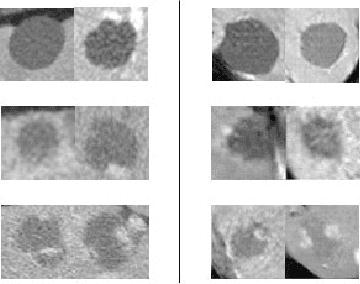
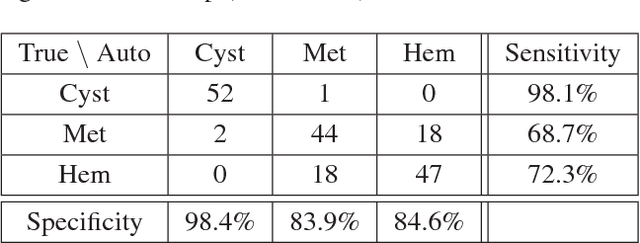
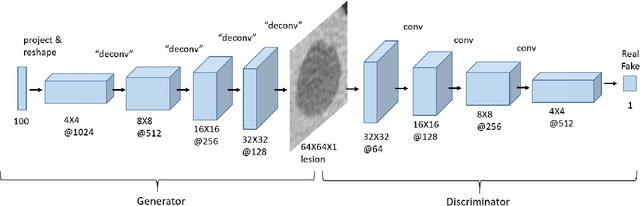
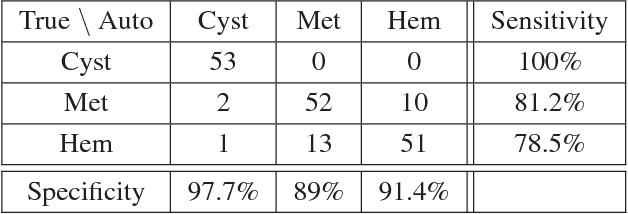
Abstract:In this paper, we present a data augmentation method that generates synthetic medical images using Generative Adversarial Networks (GANs). We propose a training scheme that first uses classical data augmentation to enlarge the training set and then further enlarges the data size and its diversity by applying GAN techniques for synthetic data augmentation. Our method is demonstrated on a limited dataset of computed tomography (CT) images of 182 liver lesions (53 cysts, 64 metastases and 65 hemangiomas). The classification performance using only classic data augmentation yielded 78.6% sensitivity and 88.4% specificity. By adding the synthetic data augmentation the results significantly increased to 85.7% sensitivity and 92.4% specificity.
Modeling the Intra-class Variability for Liver Lesion Detection using a Multi-class Patch-based CNN
Jul 20, 2017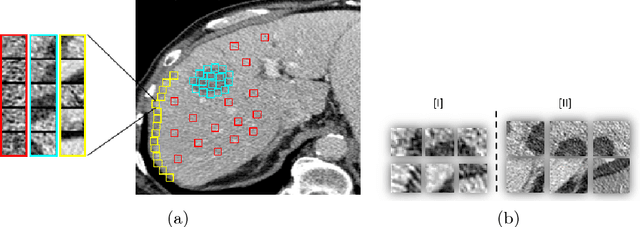
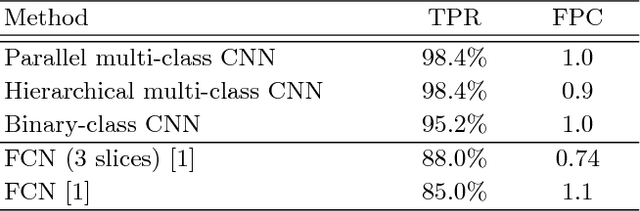
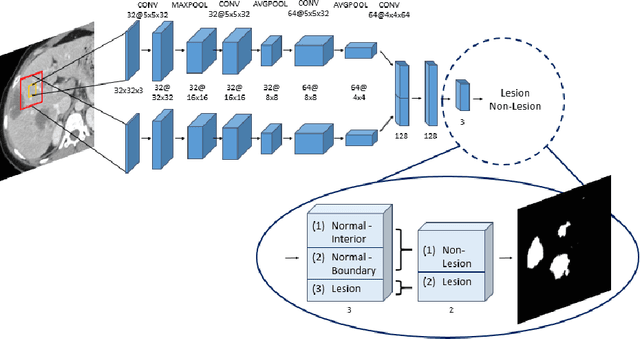
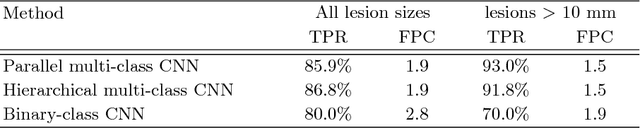
Abstract:Automatic detection of liver lesions in CT images poses a great challenge for researchers. In this work we present a deep learning approach that models explicitly the variability within the non-lesion class, based on prior knowledge of the data, to support an automated lesion detection system. A multi-class convolutional neural network (CNN) is proposed to categorize input image patches into sub-categories of boundary and interior patches, the decisions of which are fused to reach a binary lesion vs non-lesion decision. For validation of our system, we use CT images of 132 livers and 498 lesions. Our approach shows highly improved detection results that outperform the state-of-the-art fully convolutional network. Automated computerized tools, as shown in this work, have the potential in the future to support the radiologists towards improved detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge