James S. Duncan
PET Head Motion Estimation Using Supervised Deep Learning with Attention
Oct 14, 2025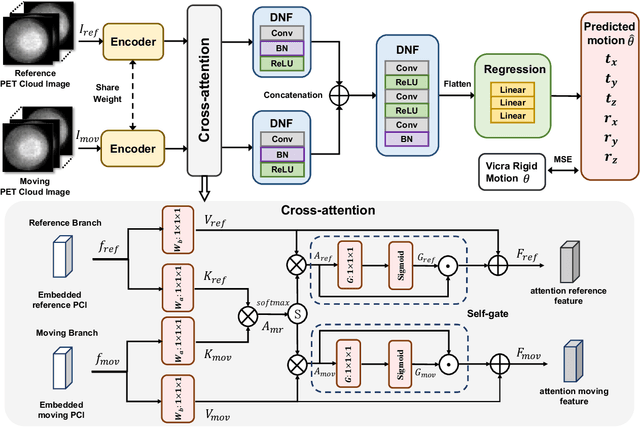
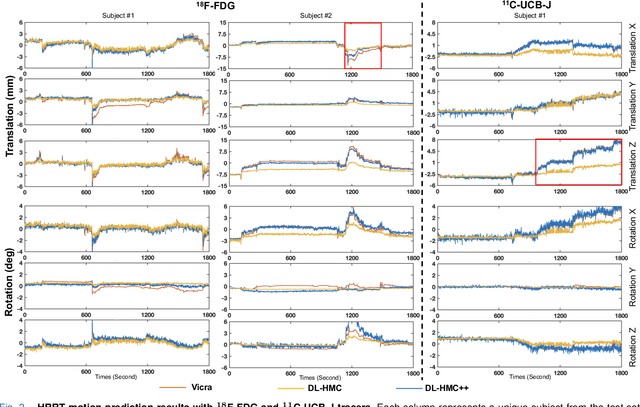
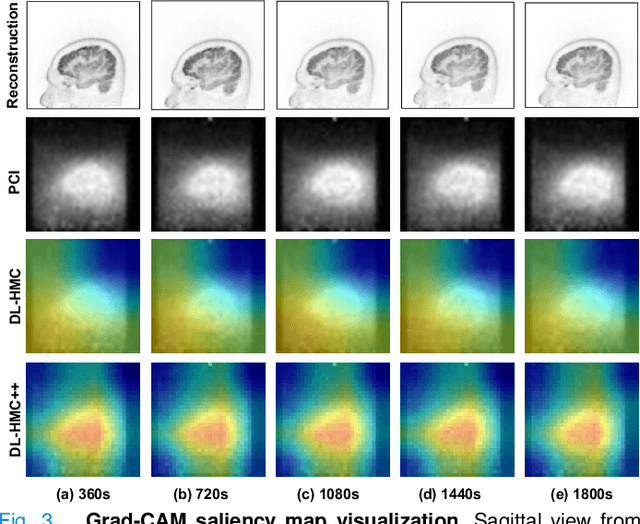
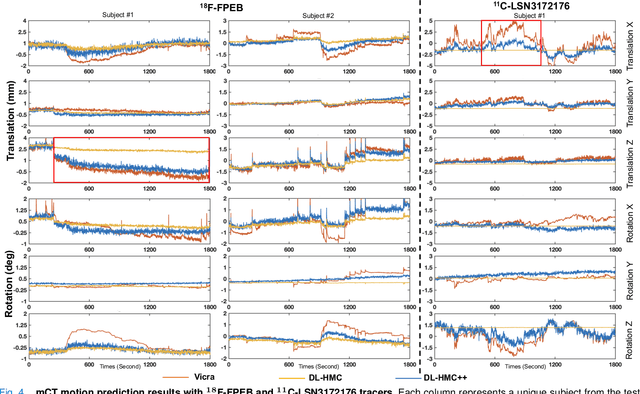
Abstract:Head movement poses a significant challenge in brain positron emission tomography (PET) imaging, resulting in image artifacts and tracer uptake quantification inaccuracies. Effective head motion estimation and correction are crucial for precise quantitative image analysis and accurate diagnosis of neurological disorders. Hardware-based motion tracking (HMT) has limited applicability in real-world clinical practice. To overcome this limitation, we propose a deep-learning head motion correction approach with cross-attention (DL-HMC++) to predict rigid head motion from one-second 3D PET raw data. DL-HMC++ is trained in a supervised manner by leveraging existing dynamic PET scans with gold-standard motion measurements from external HMT. We evaluate DL-HMC++ on two PET scanners (HRRT and mCT) and four radiotracers (18F-FDG, 18F-FPEB, 11C-UCB-J, and 11C-LSN3172176) to demonstrate the effectiveness and generalization of the approach in large cohort PET studies. Quantitative and qualitative results demonstrate that DL-HMC++ consistently outperforms state-of-the-art data-driven motion estimation methods, producing motion-free images with clear delineation of brain structures and reduced motion artifacts that are indistinguishable from gold-standard HMT. Brain region of interest standard uptake value analysis exhibits average difference ratios between DL-HMC++ and gold-standard HMT to be 1.2 plus-minus 0.5% for HRRT and 0.5 plus-minus 0.2% for mCT. DL-HMC++ demonstrates the potential for data-driven PET head motion correction to remove the burden of HMT, making motion correction accessible to clinical populations beyond research settings. The code is available at https://github.com/maxxxxxxcai/DL-HMC-TMI.
Snap-and-tune: combining deep learning and test-time optimization for high-fidelity cardiovascular volumetric meshing
Jun 09, 2025Abstract:High-quality volumetric meshing from medical images is a key bottleneck for physics-based simulations in personalized medicine. For volumetric meshing of complex medical structures, recent studies have often utilized deep learning (DL)-based template deformation approaches to enable fast test-time generation with high spatial accuracy. However, these approaches still exhibit limitations, such as limited flexibility at high-curvature areas and unrealistic inter-part distances. In this study, we introduce a simple yet effective snap-and-tune strategy that sequentially applies DL and test-time optimization, which combines fast initial shape fitting with more detailed sample-specific mesh corrections. Our method provides significant improvements in both spatial accuracy and mesh quality, while being fully automated and requiring no additional training labels. Finally, we demonstrate the versatility and usefulness of our newly generated meshes via solid mechanics simulations in two different software platforms. Our code is available at https://github.com/danpak94/Deep-Cardiac-Volumetric-Mesh.
Using Foundation Models as Pseudo-Label Generators for Pre-Clinical 4D Cardiac CT Segmentation
May 14, 2025Abstract:Cardiac image segmentation is an important step in many cardiac image analysis and modeling tasks such as motion tracking or simulations of cardiac mechanics. While deep learning has greatly advanced segmentation in clinical settings, there is limited work on pre-clinical imaging, notably in porcine models, which are often used due to their anatomical and physiological similarity to humans. However, differences between species create a domain shift that complicates direct model transfer from human to pig data. Recently, foundation models trained on large human datasets have shown promise for robust medical image segmentation; yet their applicability to porcine data remains largely unexplored. In this work, we investigate whether foundation models can generate sufficiently accurate pseudo-labels for pig cardiac CT and propose a simple self-training approach to iteratively refine these labels. Our method requires no manually annotated pig data, relying instead on iterative updates to improve segmentation quality. We demonstrate that this self-training process not only enhances segmentation accuracy but also smooths out temporal inconsistencies across consecutive frames. Although our results are encouraging, there remains room for improvement, for example by incorporating more sophisticated self-training strategies and by exploring additional foundation models and other cardiac imaging technologies.
Adapting Vision Foundation Models for Real-time Ultrasound Image Segmentation
Mar 31, 2025Abstract:We propose a novel approach that adapts hierarchical vision foundation models for real-time ultrasound image segmentation. Existing ultrasound segmentation methods often struggle with adaptability to new tasks, relying on costly manual annotations, while real-time approaches generally fail to match state-of-the-art performance. To overcome these limitations, we introduce an adaptive framework that leverages the vision foundation model Hiera to extract multi-scale features, interleaved with DINOv2 representations to enhance visual expressiveness. These enriched features are then decoded to produce precise and robust segmentation. We conduct extensive evaluations on six public datasets and one in-house dataset, covering both cardiac and thyroid ultrasound segmentation. Experiments show that our approach outperforms state-of-the-art methods across multiple datasets and excels with limited supervision, surpassing nnUNet by over 20\% on average in the 1\% and 10\% data settings. Our method achieves $\sim$77 FPS inference speed with TensorRT on a single GPU, enabling real-time clinical applications.
Progressive Test Time Energy Adaptation for Medical Image Segmentation
Mar 20, 2025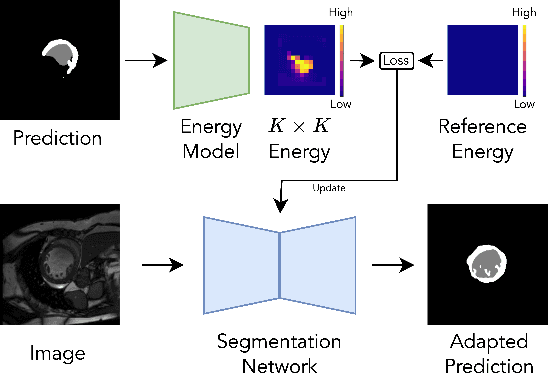
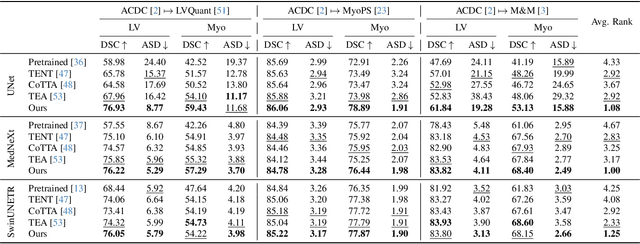
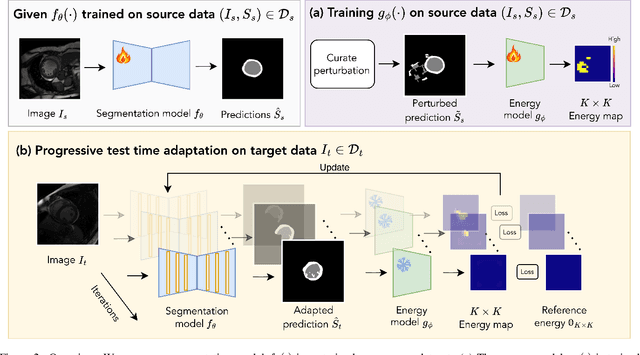
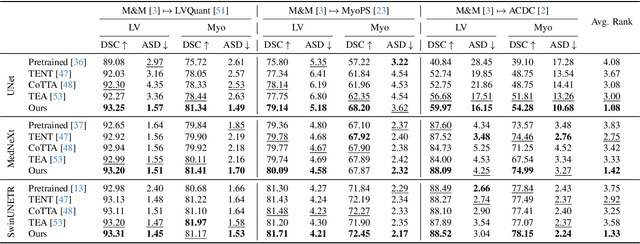
Abstract:We propose a model-agnostic, progressive test-time energy adaptation approach for medical image segmentation. Maintaining model performance across diverse medical datasets is challenging, as distribution shifts arise from inconsistent imaging protocols and patient variations. Unlike domain adaptation methods that require multiple passes through target data - impractical in clinical settings - our approach adapts pretrained models progressively as they process test data. Our method leverages a shape energy model trained on source data, which assigns an energy score at the patch level to segmentation maps: low energy represents in-distribution (accurate) shapes, while high energy signals out-of-distribution (erroneous) predictions. By minimizing this energy score at test time, we refine the segmentation model to align with the target distribution. To validate the effectiveness and adaptability, we evaluated our framework on eight public MRI (bSSFP, T1- and T2-weighted) and X-ray datasets spanning cardiac, spinal cord, and lung segmentation. We consistently outperform baselines both quantitatively and qualitatively.
Causal Modeling of fMRI Time-series for Interpretable Autism Spectrum Disorder Classification
Feb 21, 2025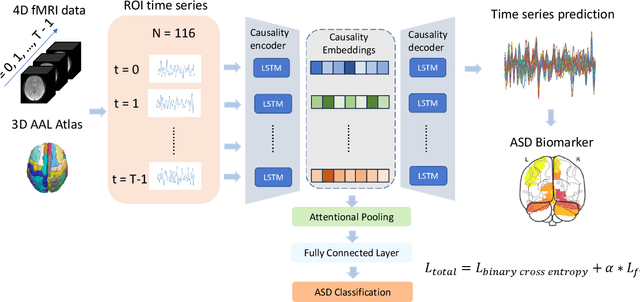
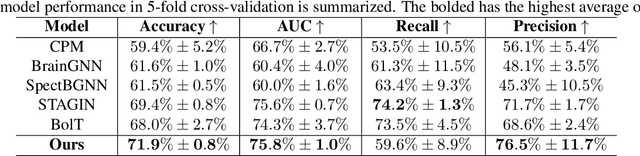
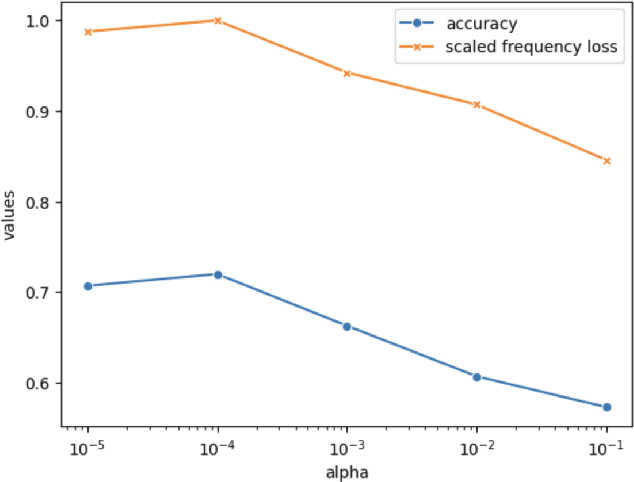
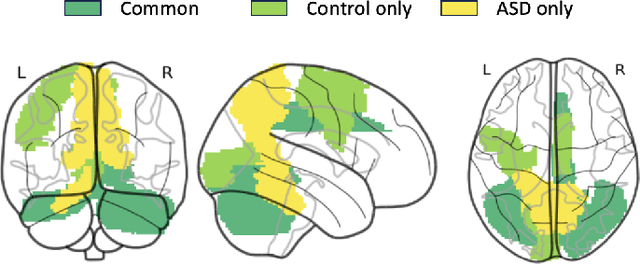
Abstract:Autism spectrum disorder (ASD) is a neurological and developmental disorder that affects social and communicative behaviors. It emerges in early life and is generally associated with lifelong disabilities. Thus, accurate and early diagnosis could facilitate treatment outcomes for those with ASD. Functional magnetic resonance imaging (fMRI) is a useful tool that measures changes in brain signaling to facilitate our understanding of ASD. Much effort is being made to identify ASD biomarkers using various connectome-based machine learning and deep learning classifiers. However, correlation-based models cannot capture the non-linear interactions between brain regions. To solve this problem, we introduce a causality-inspired deep learning model that uses time-series information from fMRI and captures causality among ROIs useful for ASD classification. The model is compared with other baseline and state-of-the-art models with 5-fold cross-validation on the ABIDE dataset. We filtered the dataset by choosing all the images with mean FD less than 15mm to ensure data quality. Our proposed model achieved the highest average classification accuracy of 71.9% and an average AUC of 75.8%. Moreover, the inter-ROI causality interpretation of the model suggests that the left precuneus, right precuneus, and cerebellum are placed in the top 10 ROIs in inter-ROI causality among the ASD population. In contrast, these ROIs are not ranked in the top 10 in the control population. We have validated our findings with the literature and found that abnormalities in these ROIs are often associated with ASD.
The Silent Majority: Demystifying Memorization Effect in the Presence of Spurious Correlations
Jan 01, 2025Abstract:Machine learning models often rely on simple spurious features -- patterns in training data that correlate with targets but are not causally related to them, like image backgrounds in foreground classification. This reliance typically leads to imbalanced test performance across minority and majority groups. In this work, we take a closer look at the fundamental cause of such imbalanced performance through the lens of memorization, which refers to the ability to predict accurately on \textit{atypical} examples (minority groups) in the training set but failing in achieving the same accuracy in the testing set. This paper systematically shows the ubiquitous existence of spurious features in a small set of neurons within the network, providing the first-ever evidence that memorization may contribute to imbalanced group performance. Through three experimental sources of converging empirical evidence, we find the property of a small subset of neurons or channels in memorizing minority group information. Inspired by these findings, we articulate the hypothesis: the imbalanced group performance is a byproduct of ``noisy'' spurious memorization confined to a small set of neurons. To further substantiate this hypothesis, we show that eliminating these unnecessary spurious memorization patterns via a novel framework during training can significantly affect the model performance on minority groups. Our experimental results across various architectures and benchmarks offer new insights on how neural networks encode core and spurious knowledge, laying the groundwork for future research in demystifying robustness to spurious correlation.
A Flow-based Truncated Denoising Diffusion Model for Super-resolution Magnetic Resonance Spectroscopic Imaging
Oct 25, 2024Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a non-invasive imaging technique for studying metabolism and has become a crucial tool for understanding neurological diseases, cancers and diabetes. High spatial resolution MRSI is needed to characterize lesions, but in practice MRSI is acquired at low resolution due to time and sensitivity restrictions caused by the low metabolite concentrations. Therefore, there is an imperative need for a post-processing approach to generate high-resolution MRSI from low-resolution data that can be acquired fast and with high sensitivity. Deep learning-based super-resolution methods provided promising results for improving the spatial resolution of MRSI, but they still have limited capability to generate accurate and high-quality images. Recently, diffusion models have demonstrated superior learning capability than other generative models in various tasks, but sampling from diffusion models requires iterating through a large number of diffusion steps, which is time-consuming. This work introduces a Flow-based Truncated Denoising Diffusion Model (FTDDM) for super-resolution MRSI, which shortens the diffusion process by truncating the diffusion chain, and the truncated steps are estimated using a normalizing flow-based network. The network is conditioned on upscaling factors to enable multi-scale super-resolution. To train and evaluate the deep learning models, we developed a 1H-MRSI dataset acquired from 25 high-grade glioma patients. We demonstrate that FTDDM outperforms existing generative models while speeding up the sampling process by over 9-fold compared to the baseline diffusion model. Neuroradiologists' evaluations confirmed the clinical advantages of our method, which also supports uncertainty estimation and sharpness adjustment, extending its potential clinical applications.
* Accepted by Medical Image Analysis (MedIA)
AI-powered multimodal modeling of personalized hemodynamics in aortic stenosis
Jun 29, 2024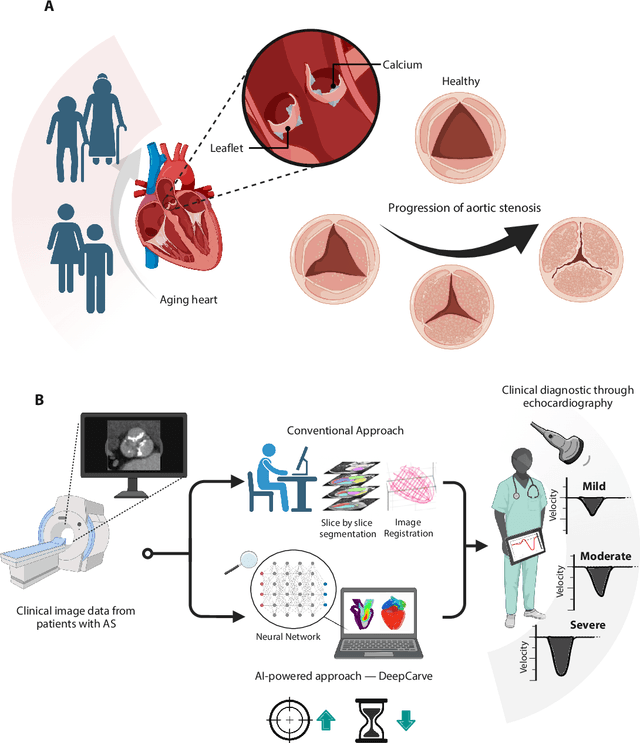
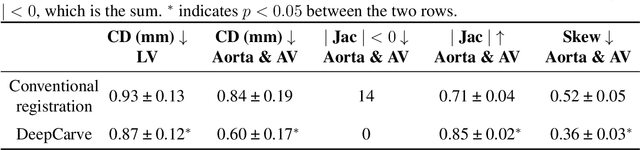
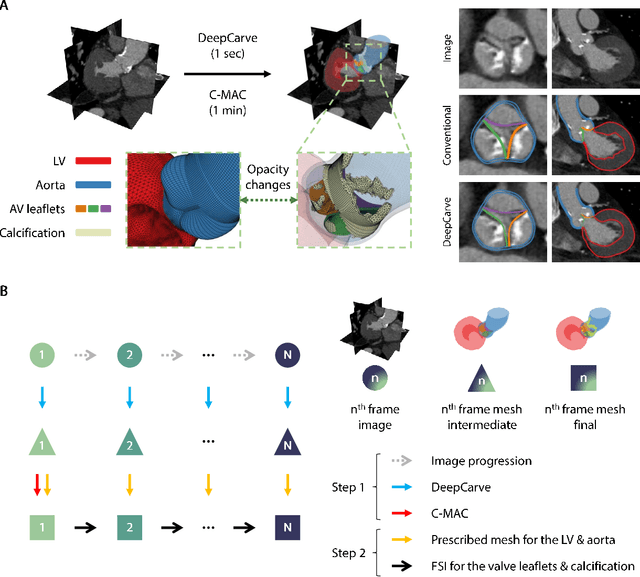
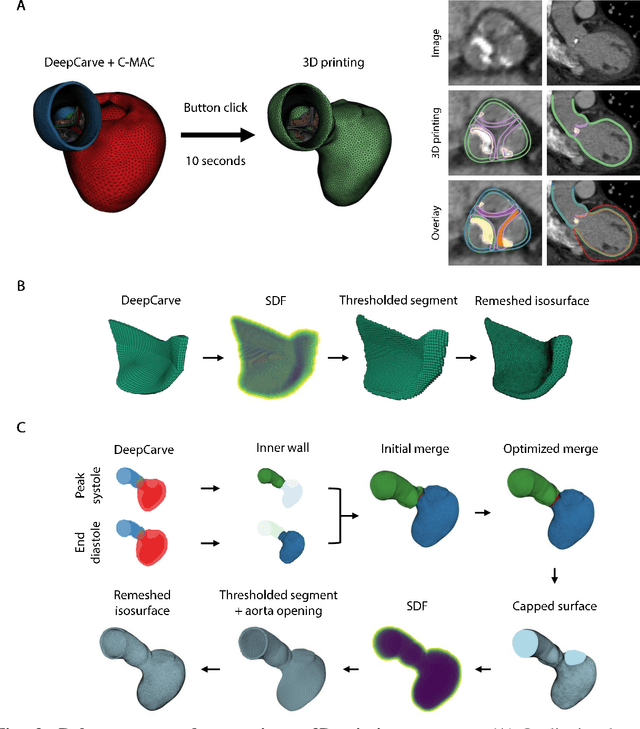
Abstract:Aortic stenosis (AS) is the most common valvular heart disease in developed countries. High-fidelity preclinical models can improve AS management by enabling therapeutic innovation, early diagnosis, and tailored treatment planning. However, their use is currently limited by complex workflows necessitating lengthy expert-driven manual operations. Here, we propose an AI-powered computational framework for accelerated and democratized patient-specific modeling of AS hemodynamics from computed tomography. First, we demonstrate that our automated meshing algorithms can generate task-ready geometries for both computational and benchtop simulations with higher accuracy and 100 times faster than existing approaches. Then, we show that our approach can be integrated with fluid-structure interaction and soft robotics models to accurately recapitulate a broad spectrum of clinical hemodynamic measurements of diverse AS patients. The efficiency and reliability of these algorithms make them an ideal complementary tool for personalized high-fidelity modeling of AS biomechanics, hemodynamics, and treatment planning.
Assessment of Clonal Hematopoiesis of Indeterminate Potential from Cardiac Magnetic Resonance Imaging using Deep Learning in a Cardio-oncology Population
Jun 26, 2024Abstract:Background: We propose a novel method to identify who may likely have clonal hematopoiesis of indeterminate potential (CHIP), a condition characterized by the presence of somatic mutations in hematopoietic stem cells without detectable hematologic malignancy, using deep learning techniques. Methods: We developed a convolutional neural network (CNN) to predict CHIP status using 4 different views from standard delayed gadolinium-enhanced cardiac magnetic resonance imaging (CMR). We used 5-fold cross validation on 82 cardio-oncology patients to assess the performance of our model. Different algorithms were compared to find the optimal patient-level prediction method using the image-level CNN predictions. Results: We found that the best model had an area under the receiver operating characteristic curve of 0.85 and an accuracy of 82%. Conclusions: We conclude that a deep learning-based diagnostic approach for CHIP using CMR is promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge