Lawrence H. Staib
Progressive Test Time Energy Adaptation for Medical Image Segmentation
Mar 20, 2025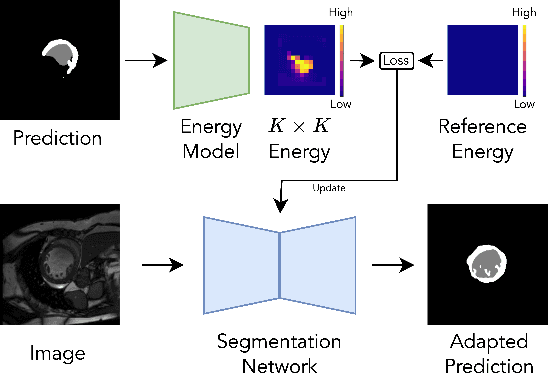
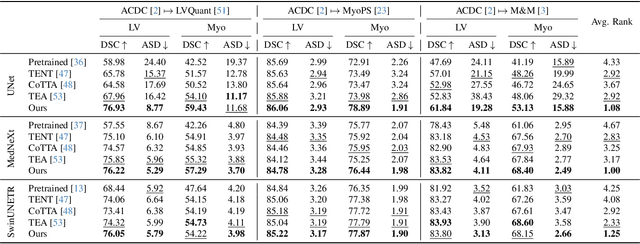
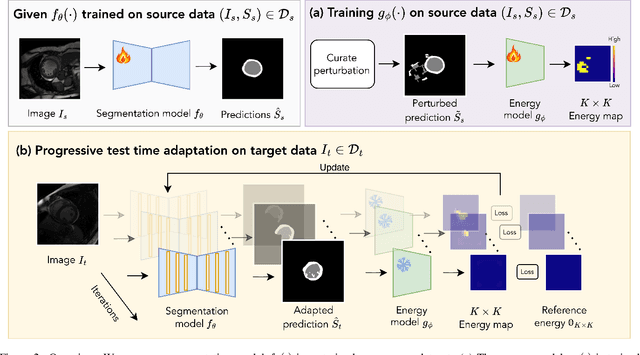
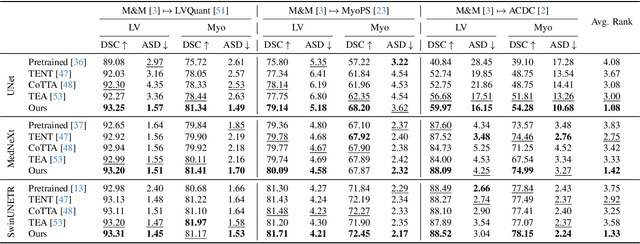
Abstract:We propose a model-agnostic, progressive test-time energy adaptation approach for medical image segmentation. Maintaining model performance across diverse medical datasets is challenging, as distribution shifts arise from inconsistent imaging protocols and patient variations. Unlike domain adaptation methods that require multiple passes through target data - impractical in clinical settings - our approach adapts pretrained models progressively as they process test data. Our method leverages a shape energy model trained on source data, which assigns an energy score at the patch level to segmentation maps: low energy represents in-distribution (accurate) shapes, while high energy signals out-of-distribution (erroneous) predictions. By minimizing this energy score at test time, we refine the segmentation model to align with the target distribution. To validate the effectiveness and adaptability, we evaluated our framework on eight public MRI (bSSFP, T1- and T2-weighted) and X-ray datasets spanning cardiac, spinal cord, and lung segmentation. We consistently outperform baselines both quantitatively and qualitatively.
Causal Modeling of fMRI Time-series for Interpretable Autism Spectrum Disorder Classification
Feb 21, 2025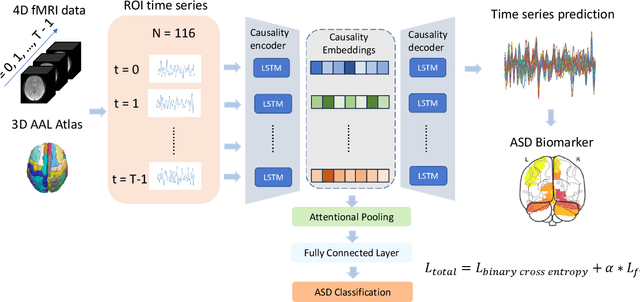
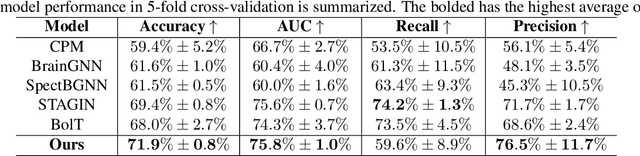
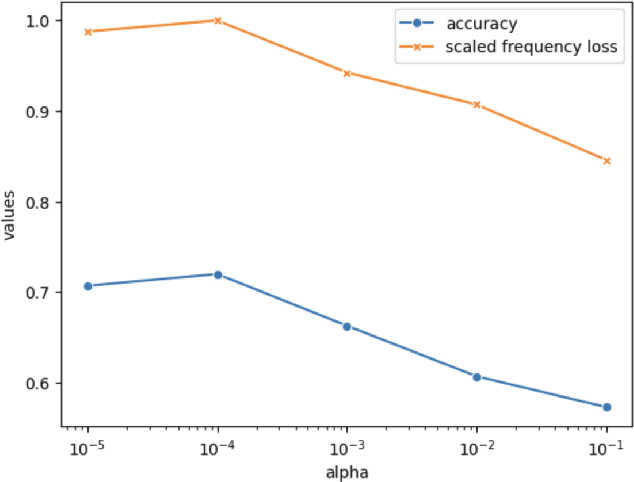
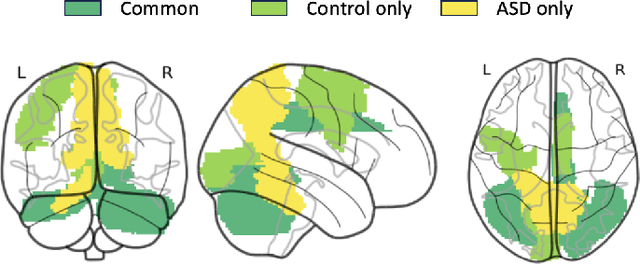
Abstract:Autism spectrum disorder (ASD) is a neurological and developmental disorder that affects social and communicative behaviors. It emerges in early life and is generally associated with lifelong disabilities. Thus, accurate and early diagnosis could facilitate treatment outcomes for those with ASD. Functional magnetic resonance imaging (fMRI) is a useful tool that measures changes in brain signaling to facilitate our understanding of ASD. Much effort is being made to identify ASD biomarkers using various connectome-based machine learning and deep learning classifiers. However, correlation-based models cannot capture the non-linear interactions between brain regions. To solve this problem, we introduce a causality-inspired deep learning model that uses time-series information from fMRI and captures causality among ROIs useful for ASD classification. The model is compared with other baseline and state-of-the-art models with 5-fold cross-validation on the ABIDE dataset. We filtered the dataset by choosing all the images with mean FD less than 15mm to ensure data quality. Our proposed model achieved the highest average classification accuracy of 71.9% and an average AUC of 75.8%. Moreover, the inter-ROI causality interpretation of the model suggests that the left precuneus, right precuneus, and cerebellum are placed in the top 10 ROIs in inter-ROI causality among the ASD population. In contrast, these ROIs are not ranked in the top 10 in the control population. We have validated our findings with the literature and found that abnormalities in these ROIs are often associated with ASD.
Enhancing Uncertainty Estimation in Semantic Segmentation via Monte-Carlo Frequency Dropout
Jan 20, 2025



Abstract:Monte-Carlo (MC) Dropout provides a practical solution for estimating predictive distributions in deterministic neural networks. Traditional dropout, applied within the signal space, may fail to account for frequency-related noise common in medical imaging, leading to biased predictive estimates. A novel approach extends Dropout to the frequency domain, allowing stochastic attenuation of signal frequencies during inference. This creates diverse global textural variations in feature maps while preserving structural integrity -- a factor we hypothesize and empirically show is contributing to accurately estimating uncertainties in semantic segmentation. We evaluated traditional MC-Dropout and the MC-frequency Dropout in three segmentation tasks involving different imaging modalities: (i) prostate zones in biparametric MRI, (ii) liver tumors in contrast-enhanced CT, and (iii) lungs in chest X-ray scans. Our results show that MC-Frequency Dropout improves calibration, convergence, and semantic uncertainty, thereby improving prediction scrutiny, boundary delineation, and has the potential to enhance medical decision-making.
Rate-In: Information-Driven Adaptive Dropout Rates for Improved Inference-Time Uncertainty Estimation
Dec 10, 2024



Abstract:Accurate uncertainty estimation is crucial for deploying neural networks in risk-sensitive applications such as medical diagnosis. Monte Carlo Dropout is a widely used technique for approximating predictive uncertainty by performing stochastic forward passes with dropout during inference. However, using static dropout rates across all layers and inputs can lead to suboptimal uncertainty estimates, as it fails to adapt to the varying characteristics of individual inputs and network layers. Existing approaches optimize dropout rates during training using labeled data, resulting in fixed inference-time parameters that cannot adjust to new data distributions, compromising uncertainty estimates in Monte Carlo simulations. In this paper, we propose Rate-In, an algorithm that dynamically adjusts dropout rates during inference by quantifying the information loss induced by dropout in each layer's feature maps. By treating dropout as controlled noise injection and leveraging information-theoretic principles, Rate-In adapts dropout rates per layer and per input instance without requiring ground truth labels. By quantifying the functional information loss in feature maps, we adaptively tune dropout rates to maintain perceptual quality across diverse medical imaging tasks and architectural configurations. Our extensive empirical study on synthetic data and real-world medical imaging tasks demonstrates that Rate-In improves calibration and sharpens uncertainty estimates compared to fixed or heuristic dropout rates without compromising predictive performance. Rate-In offers a practical, unsupervised, inference-time approach to optimizing dropout for more reliable predictive uncertainty estimation in critical applications.
STNAGNN: Spatiotemporal Node Attention Graph Neural Network for Task-based fMRI Analysis
Jun 17, 2024Abstract:Task-based fMRI uses actions or stimuli to trigger task-specific brain responses and measures them using BOLD contrast. Despite the significant task-induced spatiotemporal brain activation fluctuations, most studies on task-based fMRI ignore the task context information aligned with fMRI and consider task-based fMRI a coherent sequence. In this paper, we show that using the task structures as data-driven guidance is effective for spatiotemporal analysis. We propose STNAGNN, a GNN-based spatiotemporal architecture, and validate its performance in an autism classification task. The trained model is also interpreted for identifying autism-related spatiotemporal brain biomarkers.
TAI-GAN: A Temporally and Anatomically Informed Generative Adversarial Network for early-to-late frame conversion in dynamic cardiac PET inter-frame motion correction
Feb 14, 2024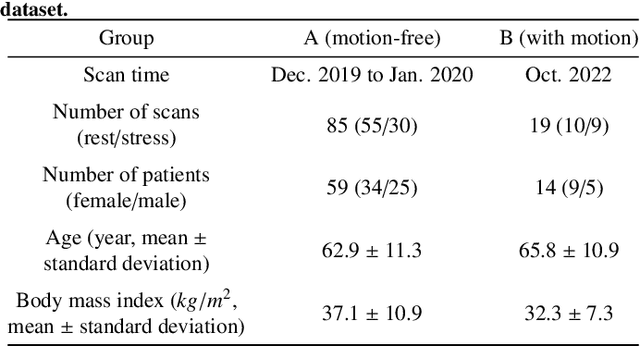
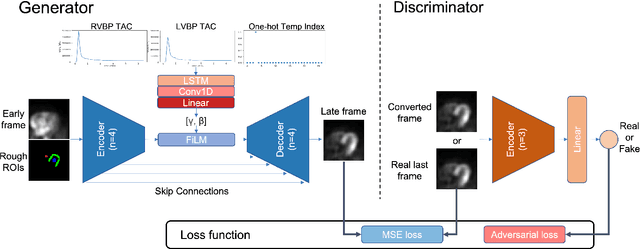
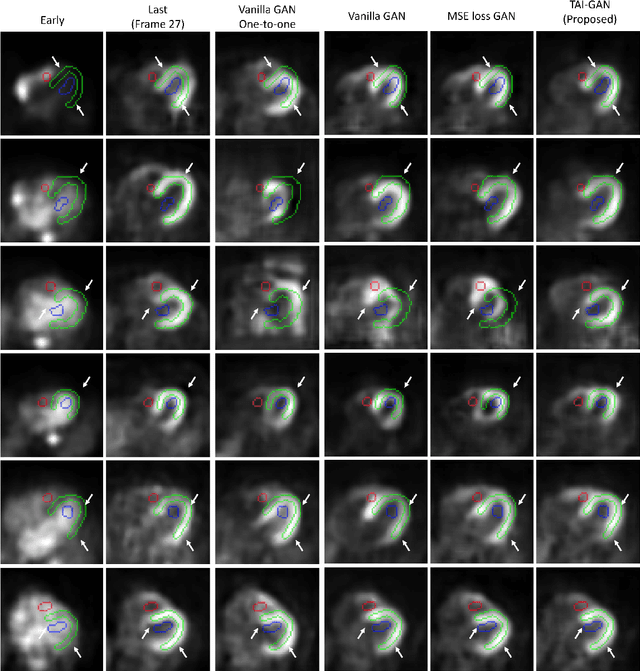
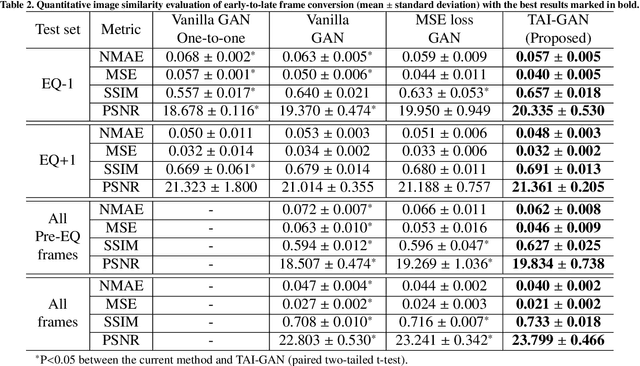
Abstract:Inter-frame motion in dynamic cardiac positron emission tomography (PET) using rubidium-82 (82-Rb) myocardial perfusion imaging impacts myocardial blood flow (MBF) quantification and the diagnosis accuracy of coronary artery diseases. However, the high cross-frame distribution variation due to rapid tracer kinetics poses a considerable challenge for inter-frame motion correction, especially for early frames where intensity-based image registration techniques often fail. To address this issue, we propose a novel method called Temporally and Anatomically Informed Generative Adversarial Network (TAI-GAN) that utilizes an all-to-one mapping to convert early frames into those with tracer distribution similar to the last reference frame. The TAI-GAN consists of a feature-wise linear modulation layer that encodes channel-wise parameters generated from temporal information and rough cardiac segmentation masks with local shifts that serve as anatomical information. Our proposed method was evaluated on a clinical 82-Rb PET dataset, and the results show that our TAI-GAN can produce converted early frames with high image quality, comparable to the real reference frames. After TAI-GAN conversion, the motion estimation accuracy and subsequent myocardial blood flow (MBF) quantification with both conventional and deep learning-based motion correction methods were improved compared to using the original frames.
Learning Sequential Information in Task-based fMRI for Synthetic Data Augmentation
Aug 29, 2023



Abstract:Insufficiency of training data is a persistent issue in medical image analysis, especially for task-based functional magnetic resonance images (fMRI) with spatio-temporal imaging data acquired using specific cognitive tasks. In this paper, we propose an approach for generating synthetic fMRI sequences that can then be used to create augmented training datasets in downstream learning tasks. To synthesize high-resolution task-specific fMRI, we adapt the $\alpha$-GAN structure, leveraging advantages of both GAN and variational autoencoder models, and propose different alternatives in aggregating temporal information. The synthetic images are evaluated from multiple perspectives including visualizations and an autism spectrum disorder (ASD) classification task. The results show that the synthetic task-based fMRI can provide effective data augmentation in learning the ASD classification task.
Learning-based Regularization for Cardiac Strain Analysis with Ability for Domain Adaptation
Jul 12, 2018



Abstract:Reliable motion estimation and strain analysis using 3D+time echocardiography (4DE) for localization and characterization of myocardial injury is valuable for early detection and targeted interventions. However, motion estimation is difficult due to the low-SNR that stems from the inherent image properties of 4DE, and intelligent regularization is critical for producing reliable motion estimates. In this work, we incorporated the notion of domain adaptation into a supervised neural network regularization framework. We first propose an unsupervised autoencoder network with biomechanical constraints for learning a latent representation that is shown to have more physiologically plausible displacements. We extended this framework to include a supervised loss term on synthetic data and showed the effects of biomechanical constraints on the network's ability for domain adaptation. We validated both the autoencoder and semi-supervised regularization method on in vivo data with implanted sonomicrometers. Finally, we showed the ability of our semi-supervised learning regularization approach to identify infarcted regions using estimated regional strain maps with good agreement to manually traced infarct regions from postmortem excised hearts.
Prediction of Autism Treatment Response from Baseline fMRI using Random Forests and Tree Bagging
May 24, 2018



Abstract:Treating children with autism spectrum disorders (ASD) with behavioral interventions, such as Pivotal Response Treatment (PRT), has shown promise in recent studies. However, deciding which therapy is best for a given patient is largely by trial and error, and choosing an ineffective intervention results in loss of valuable treatment time. We propose predicting patient response to PRT from baseline task-based fMRI by the novel application of a random forest and tree bagging strategy. Our proposed learning pipeline uses random forest regression to determine candidate brain voxels that may be informative in predicting treatment response. The candidate voxels are then tested stepwise for inclusion in a bagged tree ensemble. After the predictive model is constructed, bias correction is performed to further increase prediction accuracy. Using data from 19 ASD children who underwent a 16 week trial of PRT and a leave-one-out cross-validation framework, the presented learning pipeline was tested against several standard methods and variations of the pipeline and resulted in the highest prediction accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge