Albert J. Sinusas
Using Foundation Models as Pseudo-Label Generators for Pre-Clinical 4D Cardiac CT Segmentation
May 14, 2025Abstract:Cardiac image segmentation is an important step in many cardiac image analysis and modeling tasks such as motion tracking or simulations of cardiac mechanics. While deep learning has greatly advanced segmentation in clinical settings, there is limited work on pre-clinical imaging, notably in porcine models, which are often used due to their anatomical and physiological similarity to humans. However, differences between species create a domain shift that complicates direct model transfer from human to pig data. Recently, foundation models trained on large human datasets have shown promise for robust medical image segmentation; yet their applicability to porcine data remains largely unexplored. In this work, we investigate whether foundation models can generate sufficiently accurate pseudo-labels for pig cardiac CT and propose a simple self-training approach to iteratively refine these labels. Our method requires no manually annotated pig data, relying instead on iterative updates to improve segmentation quality. We demonstrate that this self-training process not only enhances segmentation accuracy but also smooths out temporal inconsistencies across consecutive frames. Although our results are encouraging, there remains room for improvement, for example by incorporating more sophisticated self-training strategies and by exploring additional foundation models and other cardiac imaging technologies.
A Generalizable 3D Diffusion Framework for Low-Dose and Few-View Cardiac SPECT
Dec 21, 2024Abstract:Myocardial perfusion imaging using SPECT is widely utilized to diagnose coronary artery diseases, but image quality can be negatively affected in low-dose and few-view acquisition settings. Although various deep learning methods have been introduced to improve image quality from low-dose or few-view SPECT data, previous approaches often fail to generalize across different acquisition settings, limiting their applicability in reality. This work introduced DiffSPECT-3D, a diffusion framework for 3D cardiac SPECT imaging that effectively adapts to different acquisition settings without requiring further network re-training or fine-tuning. Using both image and projection data, a consistency strategy is proposed to ensure that diffusion sampling at each step aligns with the low-dose/few-view projection measurements, the image data, and the scanner geometry, thus enabling generalization to different low-dose/few-view settings. Incorporating anatomical spatial information from CT and total variation constraint, we proposed a 2.5D conditional strategy to allow the DiffSPECT-3D to observe 3D contextual information from the entire image volume, addressing the 3D memory issues in diffusion model. We extensively evaluated the proposed method on 1,325 clinical 99mTc tetrofosmin stress/rest studies from 795 patients. Each study was reconstructed into 5 different low-count and 5 different few-view levels for model evaluations, ranging from 1% to 50% and from 1 view to 9 view, respectively. Validated against cardiac catheterization results and diagnostic comments from nuclear cardiologists, the presented results show the potential to achieve low-dose and few-view SPECT imaging without compromising clinical performance. Additionally, DiffSPECT-3D could be directly applied to full-dose SPECT images to further improve image quality, especially in a low-dose stress-first cardiac SPECT imaging protocol.
Noise-aware Dynamic Image Denoising and Positron Range Correction for Rubidium-82 Cardiac PET Imaging via Self-supervision
Sep 17, 2024



Abstract:Rb-82 is a radioactive isotope widely used for cardiac PET imaging. Despite numerous benefits of 82-Rb, there are several factors that limits its image quality and quantitative accuracy. First, the short half-life of 82-Rb results in noisy dynamic frames. Low signal-to-noise ratio would result in inaccurate and biased image quantification. Noisy dynamic frames also lead to highly noisy parametric images. The noise levels also vary substantially in different dynamic frames due to radiotracer decay and short half-life. Existing denoising methods are not applicable for this task due to the lack of paired training inputs/labels and inability to generalize across varying noise levels. Second, 82-Rb emits high-energy positrons. Compared with other tracers such as 18-F, 82-Rb travels a longer distance before annihilation, which negatively affect image spatial resolution. Here, the goal of this study is to propose a self-supervised method for simultaneous (1) noise-aware dynamic image denoising and (2) positron range correction for 82-Rb cardiac PET imaging. Tested on a series of PET scans from a cohort of normal volunteers, the proposed method produced images with superior visual quality. To demonstrate the improvement in image quantification, we compared image-derived input functions (IDIFs) with arterial input functions (AIFs) from continuous arterial blood samples. The IDIF derived from the proposed method led to lower AUC differences, decreasing from 11.09% to 7.58% on average, compared to the original dynamic frames. The proposed method also improved the quantification of myocardium blood flow (MBF), as validated against 15-O-water scans, with mean MBF differences decreased from 0.43 to 0.09, compared to the original dynamic frames. We also conducted a generalizability experiment on 37 patient scans obtained from a different country using a different scanner.
TAI-GAN: A Temporally and Anatomically Informed Generative Adversarial Network for early-to-late frame conversion in dynamic cardiac PET inter-frame motion correction
Feb 14, 2024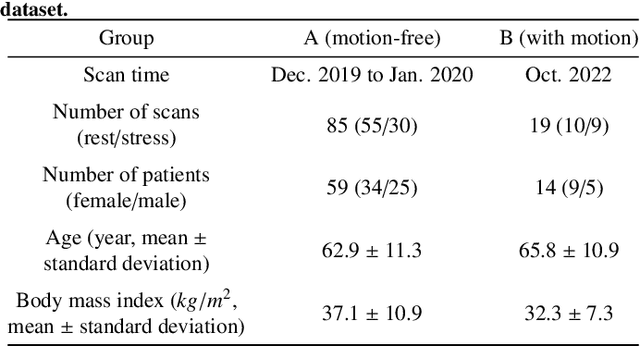
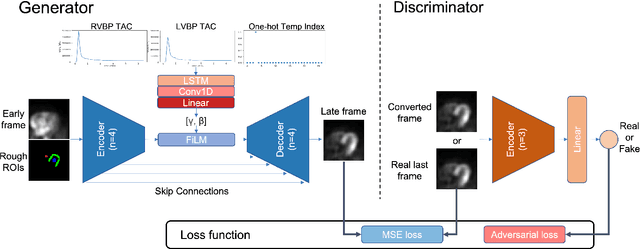
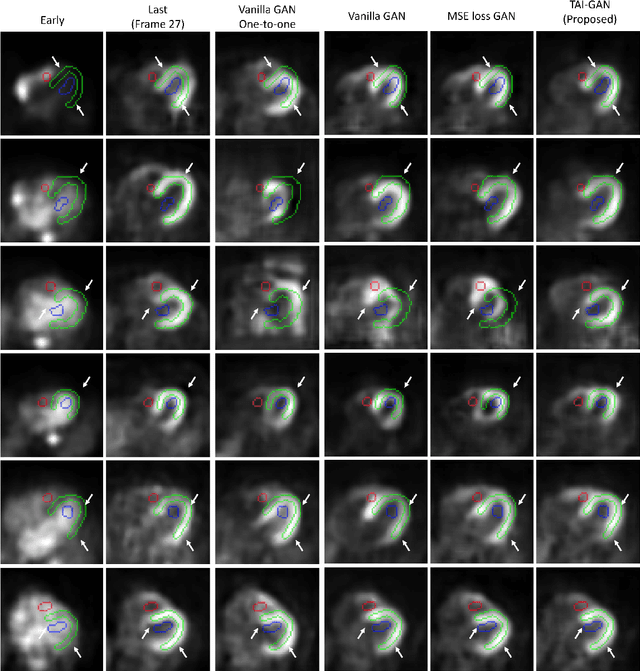
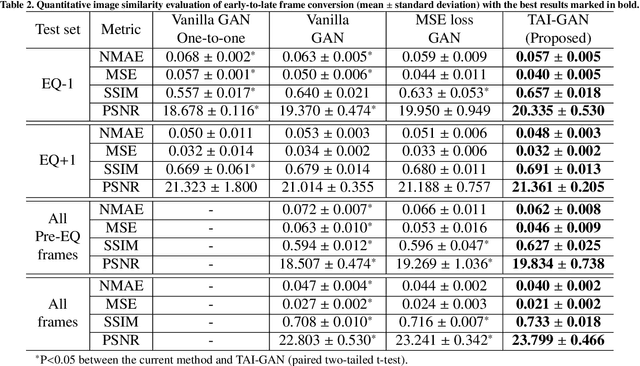
Abstract:Inter-frame motion in dynamic cardiac positron emission tomography (PET) using rubidium-82 (82-Rb) myocardial perfusion imaging impacts myocardial blood flow (MBF) quantification and the diagnosis accuracy of coronary artery diseases. However, the high cross-frame distribution variation due to rapid tracer kinetics poses a considerable challenge for inter-frame motion correction, especially for early frames where intensity-based image registration techniques often fail. To address this issue, we propose a novel method called Temporally and Anatomically Informed Generative Adversarial Network (TAI-GAN) that utilizes an all-to-one mapping to convert early frames into those with tracer distribution similar to the last reference frame. The TAI-GAN consists of a feature-wise linear modulation layer that encodes channel-wise parameters generated from temporal information and rough cardiac segmentation masks with local shifts that serve as anatomical information. Our proposed method was evaluated on a clinical 82-Rb PET dataset, and the results show that our TAI-GAN can produce converted early frames with high image quality, comparable to the real reference frames. After TAI-GAN conversion, the motion estimation accuracy and subsequent myocardial blood flow (MBF) quantification with both conventional and deep learning-based motion correction methods were improved compared to using the original frames.
Dual-Domain Coarse-to-Fine Progressive Estimation Network for Simultaneous Denoising, Limited-View Reconstruction, and Attenuation Correction of Cardiac SPECT
Jan 23, 2024Abstract:Single-Photon Emission Computed Tomography (SPECT) is widely applied for the diagnosis of coronary artery diseases. Low-dose (LD) SPECT aims to minimize radiation exposure but leads to increased image noise. Limited-view (LV) SPECT, such as the latest GE MyoSPECT ES system, enables accelerated scanning and reduces hardware expenses but degrades reconstruction accuracy. Additionally, Computed Tomography (CT) is commonly used to derive attenuation maps ($\mu$-maps) for attenuation correction (AC) of cardiac SPECT, but it will introduce additional radiation exposure and SPECT-CT misalignments. Although various methods have been developed to solely focus on LD denoising, LV reconstruction, or CT-free AC in SPECT, the solution for simultaneously addressing these tasks remains challenging and under-explored. Furthermore, it is essential to explore the potential of fusing cross-domain and cross-modality information across these interrelated tasks to further enhance the accuracy of each task. Thus, we propose a Dual-Domain Coarse-to-Fine Progressive Network (DuDoCFNet), a multi-task learning method for simultaneous LD denoising, LV reconstruction, and CT-free $\mu$-map generation of cardiac SPECT. Paired dual-domain networks in DuDoCFNet are cascaded using a multi-layer fusion mechanism for cross-domain and cross-modality feature fusion. Two-stage progressive learning strategies are applied in both projection and image domains to achieve coarse-to-fine estimations of SPECT projections and CT-derived $\mu$-maps. Our experiments demonstrate DuDoCFNet's superior accuracy in estimating projections, generating $\mu$-maps, and AC reconstructions compared to existing single- or multi-task learning methods, under various iterations and LD levels. The source code of this work is available at https://github.com/XiongchaoChen/DuDoCFNet-MultiTask.
An Adaptive Correspondence Scoring Framework for Unsupervised Image Registration of Medical Images
Dec 01, 2023



Abstract:We propose an adaptive training scheme for unsupervised medical image registration. Existing methods rely on image reconstruction as the primary supervision signal. However, nuisance variables (e.g. noise and covisibility) often cause the loss of correspondence between medical images, violating the Lambertian assumption in physical waves (e.g. ultrasound) and consistent imaging acquisition. As the unsupervised learning scheme relies on intensity constancy to establish correspondence between images for reconstruction, this introduces spurious error residuals that are not modeled by the typical training objective. To mitigate this, we propose an adaptive framework that re-weights the error residuals with a correspondence scoring map during training, preventing the parametric displacement estimator from drifting away due to noisy gradients, which leads to performance degradations. To illustrate the versatility and effectiveness of our method, we tested our framework on three representative registration architectures across three medical image datasets along with other baselines. Our proposed adaptive framework consistently outperforms other methods both quantitatively and qualitatively. Paired t-tests show that our improvements are statistically significant. The code will be publicly available at \url{https://voldemort108x.github.io/AdaCS/}.
Heteroscedastic Uncertainty Estimation for Probabilistic Unsupervised Registration of Noisy Medical Images
Dec 01, 2023



Abstract:This paper proposes a heteroscedastic uncertainty estimation framework for unsupervised medical image registration. Existing methods rely on objectives (e.g. mean-squared error) that assume a uniform noise level across the image, disregarding the heteroscedastic and input-dependent characteristics of noise distribution in real-world medical images. This further introduces noisy gradients due to undesired penalization on outliers, causing unnatural deformation and performance degradation. To mitigate this, we propose an adaptive weighting scheme with a relative $\gamma$-exponentiated signal-to-noise ratio (SNR) for the displacement estimator after modeling the heteroscedastic noise using a separate variance estimator to prevent the model from being driven away by spurious gradients from error residuals, leading to more accurate displacement estimation. To illustrate the versatility and effectiveness of the proposed method, we tested our framework on two representative registration architectures across three medical image datasets. Our proposed framework consistently outperforms other baselines both quantitatively and qualitatively while also providing accurate and sensible uncertainty measures. Paired t-tests show that our improvements in registration accuracy are statistically significant. The code will be publicly available at \url{https://voldemort108x.github.io/hetero_uncertainty/}.
TAI-GAN: Temporally and Anatomically Informed GAN for early-to-late frame conversion in dynamic cardiac PET motion correction
Aug 23, 2023Abstract:The rapid tracer kinetics of rubidium-82 ($^{82}$Rb) and high variation of cross-frame distribution in dynamic cardiac positron emission tomography (PET) raise significant challenges for inter-frame motion correction, particularly for the early frames where conventional intensity-based image registration techniques are not applicable. Alternatively, a promising approach utilizes generative methods to handle the tracer distribution changes to assist existing registration methods. To improve frame-wise registration and parametric quantification, we propose a Temporally and Anatomically Informed Generative Adversarial Network (TAI-GAN) to transform the early frames into the late reference frame using an all-to-one mapping. Specifically, a feature-wise linear modulation layer encodes channel-wise parameters generated from temporal tracer kinetics information, and rough cardiac segmentations with local shifts serve as the anatomical information. We validated our proposed method on a clinical $^{82}$Rb PET dataset and found that our TAI-GAN can produce converted early frames with high image quality, comparable to the real reference frames. After TAI-GAN conversion, motion estimation accuracy and clinical myocardial blood flow (MBF) quantification were improved compared to using the original frames. Our code is published at https://github.com/gxq1998/TAI-GAN.
Joint Denoising and Few-angle Reconstruction for Low-dose Cardiac SPECT Using a Dual-domain Iterative Network with Adaptive Data Consistency
May 17, 2023



Abstract:Myocardial perfusion imaging (MPI) by single-photon emission computed tomography (SPECT) is widely applied for the diagnosis of cardiovascular diseases. Reducing the dose of the injected tracer is essential for lowering the patient's radiation exposure, but it will lead to increased image noise. Additionally, the latest dedicated cardiac SPECT scanners typically acquire projections in fewer angles using fewer detectors to reduce hardware expenses, potentially resulting in lower reconstruction accuracy. To overcome these challenges, we propose a dual-domain iterative network for end-to-end joint denoising and reconstruction from low-dose and few-angle projections of cardiac SPECT. The image-domain network provides a prior estimate for the projection-domain networks. The projection-domain primary and auxiliary modules are interconnected for progressive denoising and few-angle reconstruction. Adaptive Data Consistency (ADC) modules improve prediction accuracy by efficiently fusing the outputs of the primary and auxiliary modules. Experiments using clinical MPI data show that our proposed method outperforms existing image-, projection-, and dual-domain techniques, producing more accurate projections and reconstructions. Ablation studies confirm the significance of the image-domain prior estimate and ADC modules in enhancing network performance.
Cross-domain Iterative Network for Simultaneous Denoising, Limited-angle Reconstruction, and Attenuation Correction of Low-dose Cardiac SPECT
May 17, 2023



Abstract:Single-Photon Emission Computed Tomography (SPECT) is widely applied for the diagnosis of ischemic heart diseases. Low-dose (LD) SPECT aims to minimize radiation exposure but leads to increased image noise. Limited-angle (LA) SPECT enables faster scanning and reduced hardware costs but results in lower reconstruction accuracy. Additionally, computed tomography (CT)-derived attenuation maps ($\mu$-maps) are commonly used for SPECT attenuation correction (AC), but it will cause extra radiation exposure and SPECT-CT misalignments. In addition, the majority of SPECT scanners in the market are not hybrid SPECT/CT scanners. Although various deep learning methods have been introduced to separately address these limitations, the solution for simultaneously addressing these challenges still remains highly under-explored and challenging. To this end, we propose a Cross-domain Iterative Network (CDI-Net) for simultaneous denoising, LA reconstruction, and CT-free AC in cardiac SPECT. In CDI-Net, paired projection- and image-domain networks are end-to-end connected to fuse the emission and anatomical information across domains and iterations. Adaptive Weight Recalibrators (AWR) adjust the multi-channel input features to enhance prediction accuracy. Our experiments using clinical data showed that CDI-Net produced more accurate $\mu$-maps, projections, and reconstructions compared to existing approaches that addressed each task separately. Ablation studies demonstrated the significance of cross-domain and cross-iteration connections, as well as AWR, in improving the reconstruction performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge