Lawrence Staib
Calibrating Multi-modal Representations: A Pursuit of Group Robustness without Annotations
Mar 12, 2024
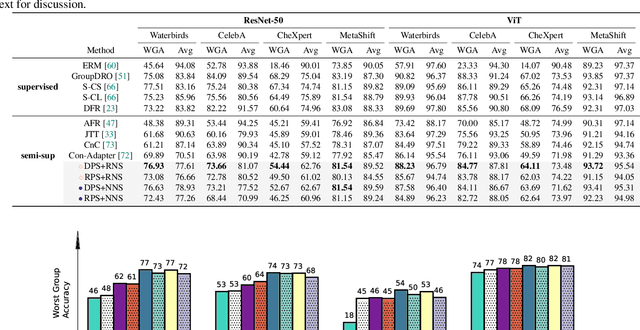

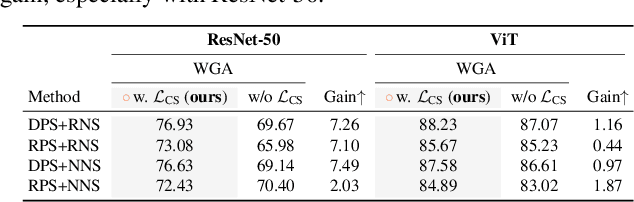
Abstract:Fine-tuning pre-trained vision-language models, like CLIP, has yielded success on diverse downstream tasks. However, several pain points persist for this paradigm: (i) directly tuning entire pre-trained models becomes both time-intensive and computationally costly. Additionally, these tuned models tend to become highly specialized, limiting their practicality for real-world deployment; (ii) recent studies indicate that pre-trained vision-language classifiers may overly depend on spurious features -- patterns that correlate with the target in training data, but are not related to the true labeling function; and (iii) existing studies on mitigating the reliance on spurious features, largely based on the assumption that we can identify such features, does not provide definitive assurance for real-world applications. As a piloting study, this work focuses on exploring mitigating the reliance on spurious features for CLIP without using any group annotation. To this end, we systematically study the existence of spurious correlation on CLIP and CILP+ERM. We first, following recent work on Deep Feature Reweighting (DFR), verify that last-layer retraining can greatly improve group robustness on pretrained CLIP. In view of them, we advocate a lightweight representation calibration method for fine-tuning CLIP, by first generating a calibration set using the pretrained CLIP, and then calibrating representations of samples within this set through contrastive learning, all without the need for group labels. Extensive experiments and in-depth visualizations on several benchmarks validate the effectiveness of our proposals, largely reducing reliance and significantly boosting the model generalization.
Heteroscedastic Uncertainty Estimation for Probabilistic Unsupervised Registration of Noisy Medical Images
Dec 01, 2023



Abstract:This paper proposes a heteroscedastic uncertainty estimation framework for unsupervised medical image registration. Existing methods rely on objectives (e.g. mean-squared error) that assume a uniform noise level across the image, disregarding the heteroscedastic and input-dependent characteristics of noise distribution in real-world medical images. This further introduces noisy gradients due to undesired penalization on outliers, causing unnatural deformation and performance degradation. To mitigate this, we propose an adaptive weighting scheme with a relative $\gamma$-exponentiated signal-to-noise ratio (SNR) for the displacement estimator after modeling the heteroscedastic noise using a separate variance estimator to prevent the model from being driven away by spurious gradients from error residuals, leading to more accurate displacement estimation. To illustrate the versatility and effectiveness of the proposed method, we tested our framework on two representative registration architectures across three medical image datasets. Our proposed framework consistently outperforms other baselines both quantitatively and qualitatively while also providing accurate and sensible uncertainty measures. Paired t-tests show that our improvements in registration accuracy are statistically significant. The code will be publicly available at \url{https://voldemort108x.github.io/hetero_uncertainty/}.
An Adaptive Correspondence Scoring Framework for Unsupervised Image Registration of Medical Images
Dec 01, 2023



Abstract:We propose an adaptive training scheme for unsupervised medical image registration. Existing methods rely on image reconstruction as the primary supervision signal. However, nuisance variables (e.g. noise and covisibility) often cause the loss of correspondence between medical images, violating the Lambertian assumption in physical waves (e.g. ultrasound) and consistent imaging acquisition. As the unsupervised learning scheme relies on intensity constancy to establish correspondence between images for reconstruction, this introduces spurious error residuals that are not modeled by the typical training objective. To mitigate this, we propose an adaptive framework that re-weights the error residuals with a correspondence scoring map during training, preventing the parametric displacement estimator from drifting away due to noisy gradients, which leads to performance degradations. To illustrate the versatility and effectiveness of our method, we tested our framework on three representative registration architectures across three medical image datasets along with other baselines. Our proposed adaptive framework consistently outperforms other methods both quantitatively and qualitatively. Paired t-tests show that our improvements are statistically significant. The code will be publicly available at \url{https://voldemort108x.github.io/AdaCS/}.
ACTION++: Improving Semi-supervised Medical Image Segmentation with Adaptive Anatomical Contrast
Apr 07, 2023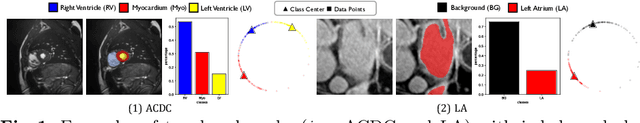
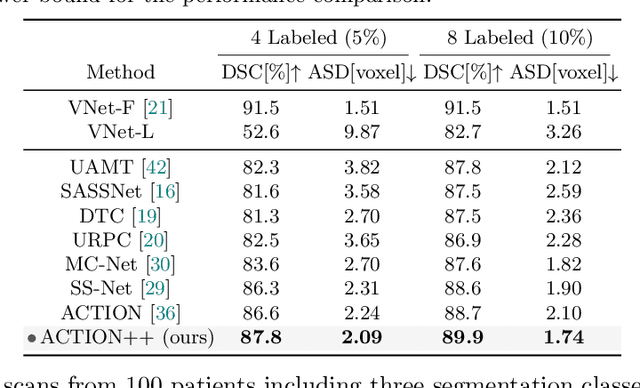
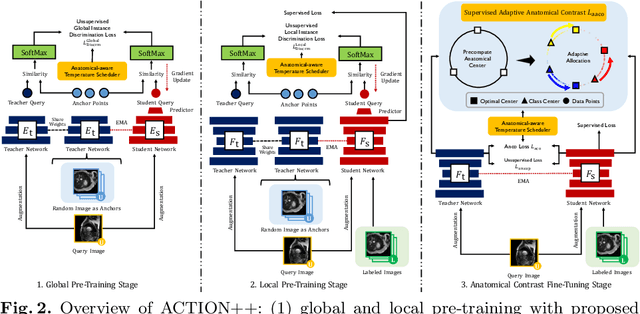
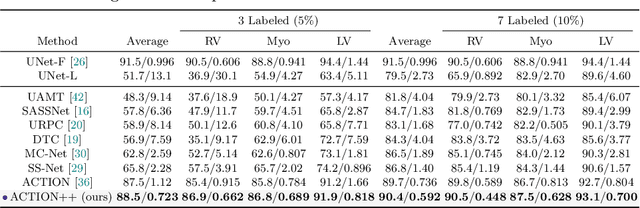
Abstract:Medical data often exhibits long-tail distributions with heavy class imbalance, which naturally leads to difficulty in classifying the minority classes (i.e., boundary regions or rare objects). Recent work has significantly improved semi-supervised medical image segmentation in long-tailed scenarios by equipping them with unsupervised contrastive criteria. However, it remains unclear how well they will perform in the labeled portion of data where class distribution is also highly imbalanced. In this work, we present ACTION++, an improved contrastive learning framework with adaptive anatomical contrast for semi-supervised medical segmentation. Specifically, we propose an adaptive supervised contrastive loss, where we first compute the optimal locations of class centers uniformly distributed on the embedding space (i.e., off-line), and then perform online contrastive matching training by encouraging different class features to adaptively match these distinct and uniformly distributed class centers. Moreover, we argue that blindly adopting a constant temperature $\tau$ in the contrastive loss on long-tailed medical data is not optimal, and propose to use a dynamic $\tau$ via a simple cosine schedule to yield better separation between majority and minority classes. Empirically, we evaluate ACTION++ on ACDC and LA benchmarks and show that it achieves state-of-the-art across two semi-supervised settings. Theoretically, we analyze the performance of adaptive anatomical contrast and confirm its superiority in label efficiency.
Implicit Anatomical Rendering for Medical Image Segmentation with Stochastic Experts
Apr 06, 2023



Abstract:Integrating high-level semantically correlated contents and low-level anatomical features is of central importance in medical image segmentation. Towards this end, recent deep learning-based medical segmentation methods have shown great promise in better modeling such information. However, convolution operators for medical segmentation typically operate on regular grids, which inherently blur the high-frequency regions, i.e., boundary regions. In this work, we propose MORSE, a generic implicit neural rendering framework designed at an anatomical level to assist learning in medical image segmentation. Our method is motivated by the fact that implicit neural representation has been shown to be more effective in fitting complex signals and solving computer graphics problems than discrete grid-based representation. The core of our approach is to formulate medical image segmentation as a rendering problem in an end-to-end manner. Specifically, we continuously align the coarse segmentation prediction with the ambiguous coordinate-based point representations and aggregate these features to adaptively refine the boundary region. To parallelly optimize multi-scale pixel-level features, we leverage the idea from Mixture-of-Expert (MoE) to design and train our MORSE with a stochastic gating mechanism. Our experiments demonstrate that MORSE can work well with different medical segmentation backbones, consistently achieving competitive performance improvements in both 2D and 3D supervised medical segmentation methods. We also theoretically analyze the superiority of MORSE.
Mine yOur owN Anatomy: Revisiting Medical Image Segmentation with Extremely Limited Labels
Sep 28, 2022



Abstract:Recent studies on contrastive learning have achieved remarkable performance solely by leveraging few labels in the context of medical image segmentation. Existing methods mainly focus on instance discrimination and invariant mapping. However, they face three common pitfalls: (1) tailness: medical image data usually follows an implicit long-tail class distribution. Blindly leveraging all pixels in training hence can lead to the data imbalance issues, and cause deteriorated performance; (2) consistency: it remains unclear whether a segmentation model has learned meaningful and yet consistent anatomical features due to the intra-class variations between different anatomical features; and (3) diversity: the intra-slice correlations within the entire dataset have received significantly less attention. This motivates us to seek a principled approach for strategically making use of the dataset itself to discover similar yet distinct samples from different anatomical views. In this paper, we introduce a novel semi-supervised medical image segmentation framework termed Mine yOur owN Anatomy (MONA), and make three contributions. First, prior work argues that every pixel equally matters to the model training; we observe empirically that this alone is unlikely to define meaningful anatomical features, mainly due to lacking the supervision signal. We show two simple solutions towards learning invariances - through the use of stronger data augmentations and nearest neighbors. Second, we construct a set of objectives that encourage the model to be capable of decomposing medical images into a collection of anatomical features in an unsupervised manner. Lastly, our extensive results on three benchmark datasets with different labeled settings validate the effectiveness of our proposed MONA which achieves new state-of-the-art under different labeled settings.
Learning correspondences of cardiac motion from images using biomechanics-informed modeling
Sep 01, 2022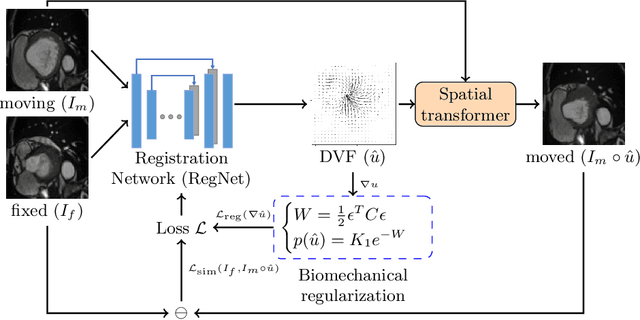
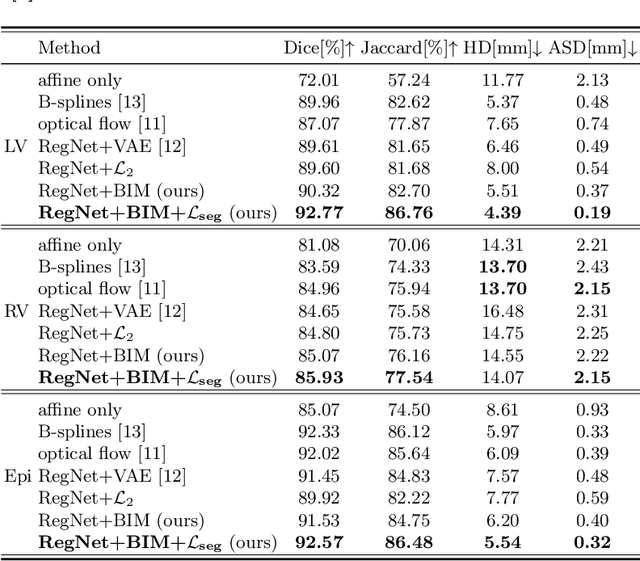
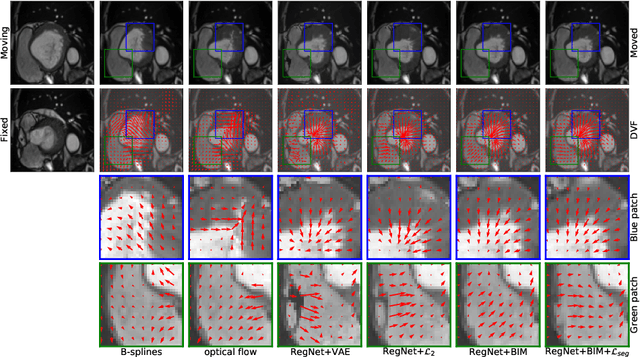
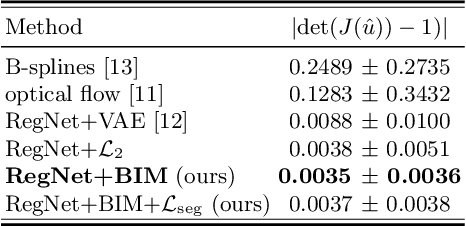
Abstract:Learning spatial-temporal correspondences in cardiac motion from images is important for understanding the underlying dynamics of cardiac anatomical structures. Many methods explicitly impose smoothness constraints such as the $\mathcal{L}_2$ norm on the displacement vector field (DVF), while usually ignoring biomechanical feasibility in the transformation. Other geometric constraints either regularize specific regions of interest such as imposing incompressibility on the myocardium or introduce additional steps such as training a separate network-based regularizer on physically simulated datasets. In this work, we propose an explicit biomechanics-informed prior as regularization on the predicted DVF in modeling a more generic biomechanically plausible transformation within all cardiac structures without introducing additional training complexity. We validate our methods on two publicly available datasets in the context of 2D MRI data and perform extensive experiments to illustrate the effectiveness and robustness of our proposed methods compared to other competing regularization schemes. Our proposed methods better preserve biomechanical properties by visual assessment and show advantages in segmentation performance using quantitative evaluation metrics. The code is publicly available at \url{https://github.com/Voldemort108X/bioinformed_reg}.
Bootstrapping Semi-supervised Medical Image Segmentation with Anatomical-aware Contrastive Distillation
Jun 06, 2022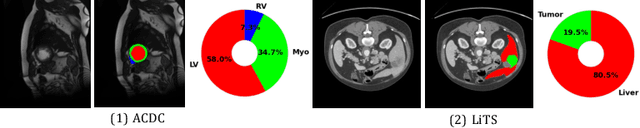

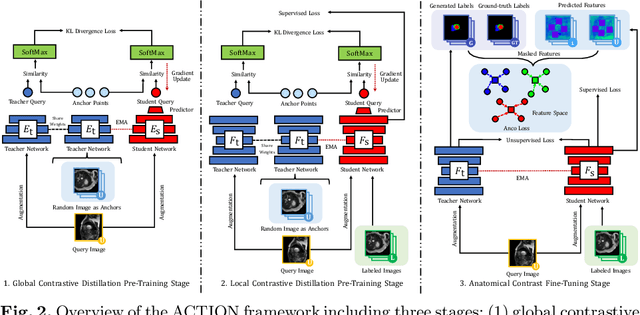
Abstract:Contrastive learning has shown great promise over annotation scarcity problems in the context of medical image segmentation. Existing approaches typically assume a balanced class distribution for both labeled and unlabeled medical images. However, medical image data in reality is commonly imbalanced (i.e., multi-class label imbalance), which naturally yields blurry contours and usually incorrectly labels rare objects. Moreover, it remains unclear whether all negative samples are equally negative. In this work, we present ACTION, an Anatomical-aware ConTrastive dIstillatiON framework, for semi-supervised medical image segmentation. Specifically, we first develop an iterative contrastive distillation algorithm by softly labeling the negatives rather than binary supervision between positive and negative pairs. We also capture more semantically similar features from the randomly chosen negative set compared to the positives to enforce the diversity of the sampled data. Second, we raise a more important question: Can we really handle imbalanced samples to yield better performance? Hence, the key innovation in ACTION is to learn global semantic relationship across the entire dataset and local anatomical features among the neighbouring pixels with minimal additional memory footprint. During the training, we introduce anatomical contrast by actively sampling a sparse set of hard negative pixels, which can generate smoother segmentation boundaries and more accurate predictions. Extensive experiments across two benchmark datasets and different unlabeled settings show that ACTION performs comparable or better than the current state-of-the-art supervised and semi-supervised methods. Our code and models will be publicly available.
Incremental Learning Meets Transfer Learning: Application to Multi-site Prostate MRI Segmentation
Jun 03, 2022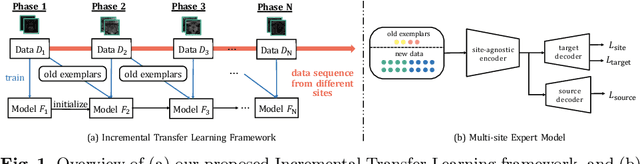

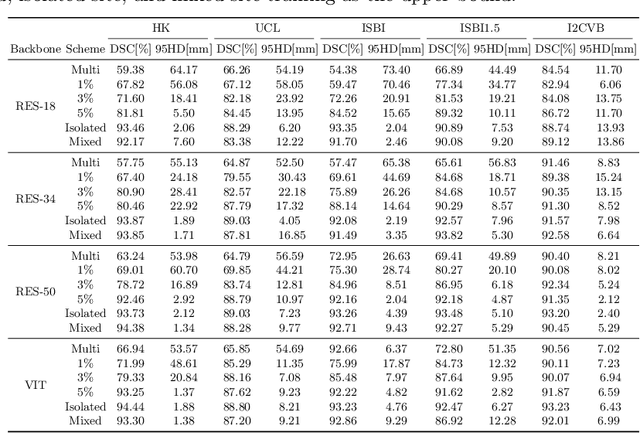
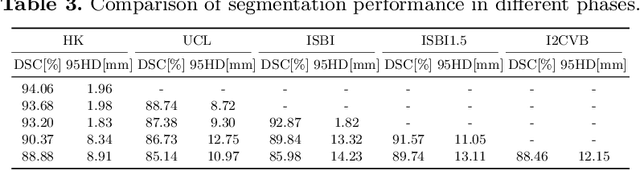
Abstract:Many medical datasets have recently been created for medical image segmentation tasks, and it is natural to question whether we can use them to sequentially train a single model that (1) performs better on all these datasets, and (2) generalizes well and transfers better to the unknown target site domain. Prior works have achieved this goal by jointly training one model on multi-site datasets, which achieve competitive performance on average but such methods rely on the assumption about the availability of all training data, thus limiting its effectiveness in practical deployment. In this paper, we propose a novel multi-site segmentation framework called incremental-transfer learning (ITL), which learns a model from multi-site datasets in an end-to-end sequential fashion. Specifically, "incremental" refers to training sequentially constructed datasets, and "transfer" is achieved by leveraging useful information from the linear combination of embedding features on each dataset. In addition, we introduce our ITL framework, where we train the network including a site-agnostic encoder with pre-trained weights and at most two segmentation decoder heads. We also design a novel site-level incremental loss in order to generalize well on the target domain. Second, we show for the first time that leveraging our ITL training scheme is able to alleviate challenging catastrophic forgetting problems in incremental learning. We conduct experiments using five challenging benchmark datasets to validate the effectiveness of our incremental-transfer learning approach. Our approach makes minimal assumptions on computation resources and domain-specific expertise, and hence constitutes a strong starting point in multi-site medical image segmentation.
Class-Aware Generative Adversarial Transformers for Medical Image Segmentation
Jan 28, 2022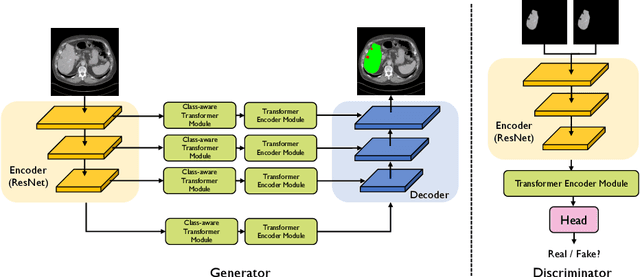
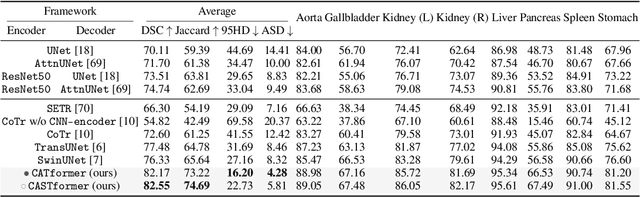
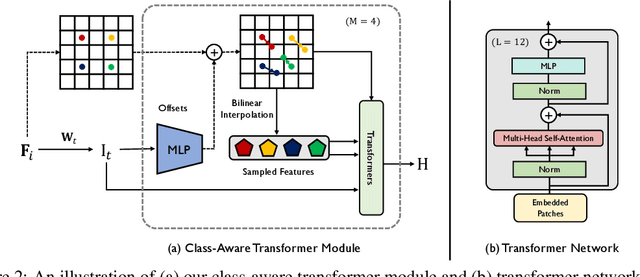
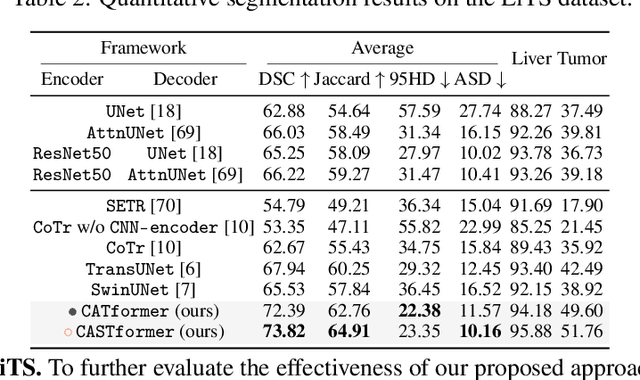
Abstract:Transformers have made remarkable progress towards modeling long-range dependencies within the medical image analysis domain. However, current transformer-based models suffer from several disadvantages: (1) existing methods fail to capture the important features of the images due to the naive tokenization scheme; (2) the models suffer from information loss because they only consider single-scale feature representations; and (3) the segmentation label maps generated by the models are not accurate enough without considering rich semantic contexts and anatomical textures. In this work, we present CA-GANformer, a novel type of generative adversarial transformers, for medical image segmentation. First, we take advantage of the pyramid structure to construct multi-scale representations and handle multi-scale variations. We then design a novel class-aware transformer module to better learn the discriminative regions of objects with semantic structures. Lastly, we utilize an adversarial training strategy that boosts segmentation accuracy and correspondingly allows a transformer-based discriminator to capture high-level semantically correlated contents and low-level anatomical features. Our experiments demonstrate that CA-GANformer dramatically outperforms previous state-of-the-art transformer-based approaches on three benchmarks, obtaining 2.54%-5.88% absolute improvements in Dice over previous models. Further qualitative experiments provide a more detailed picture of the model's inner workings, shed light on the challenges in improved transparency, and demonstrate that transfer learning can greatly improve performance and reduce the size of medical image datasets in training, making CA-GANformer a strong starting point for downstream medical image analysis tasks. Codes and models will be available to the public.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge