Shawn Ahn
Assessment of Clonal Hematopoiesis of Indeterminate Potential from Cardiac Magnetic Resonance Imaging using Deep Learning in a Cardio-oncology Population
Jun 26, 2024Abstract:Background: We propose a novel method to identify who may likely have clonal hematopoiesis of indeterminate potential (CHIP), a condition characterized by the presence of somatic mutations in hematopoietic stem cells without detectable hematologic malignancy, using deep learning techniques. Methods: We developed a convolutional neural network (CNN) to predict CHIP status using 4 different views from standard delayed gadolinium-enhanced cardiac magnetic resonance imaging (CMR). We used 5-fold cross validation on 82 cardio-oncology patients to assess the performance of our model. Different algorithms were compared to find the optimal patient-level prediction method using the image-level CNN predictions. Results: We found that the best model had an area under the receiver operating characteristic curve of 0.85 and an accuracy of 82%. Conclusions: We conclude that a deep learning-based diagnostic approach for CHIP using CMR is promising.
Learning correspondences of cardiac motion from images using biomechanics-informed modeling
Sep 01, 2022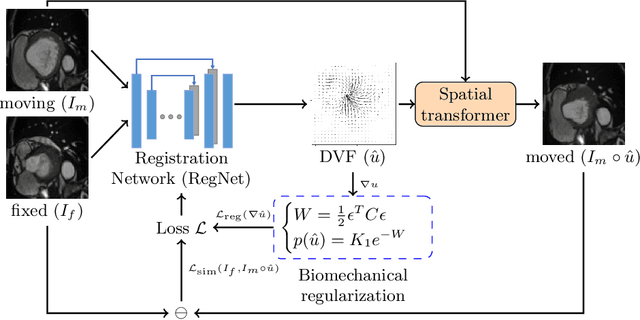
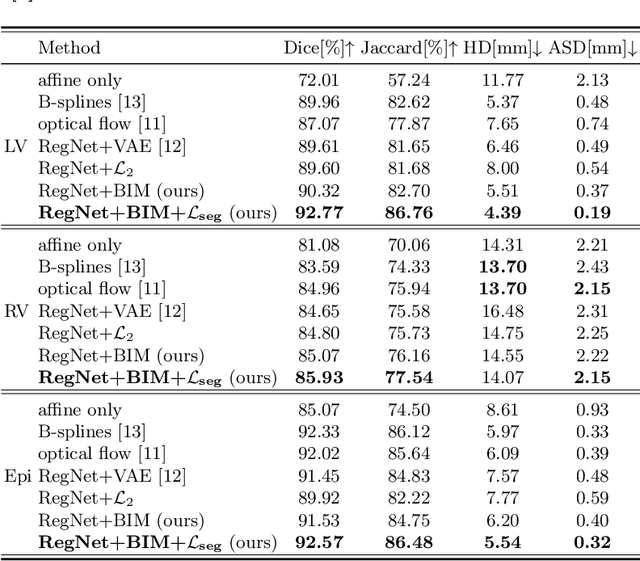
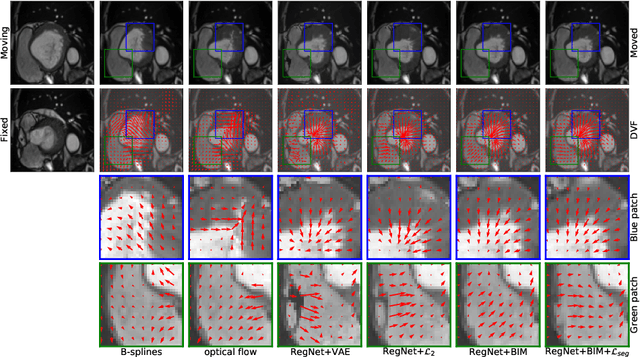
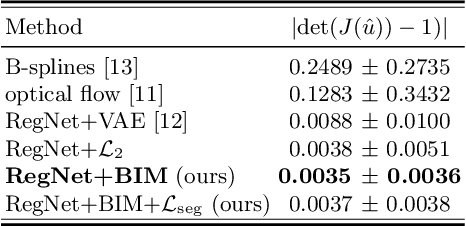
Abstract:Learning spatial-temporal correspondences in cardiac motion from images is important for understanding the underlying dynamics of cardiac anatomical structures. Many methods explicitly impose smoothness constraints such as the $\mathcal{L}_2$ norm on the displacement vector field (DVF), while usually ignoring biomechanical feasibility in the transformation. Other geometric constraints either regularize specific regions of interest such as imposing incompressibility on the myocardium or introduce additional steps such as training a separate network-based regularizer on physically simulated datasets. In this work, we propose an explicit biomechanics-informed prior as regularization on the predicted DVF in modeling a more generic biomechanically plausible transformation within all cardiac structures without introducing additional training complexity. We validate our methods on two publicly available datasets in the context of 2D MRI data and perform extensive experiments to illustrate the effectiveness and robustness of our proposed methods compared to other competing regularization schemes. Our proposed methods better preserve biomechanical properties by visual assessment and show advantages in segmentation performance using quantitative evaluation metrics. The code is publicly available at \url{https://github.com/Voldemort108X/bioinformed_reg}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge