Nicha C. Dvornek
Equi-ViT: Rotational Equivariant Vision Transformer for Robust Histopathology Analysis
Jan 14, 2026Abstract:Vision Transformers (ViTs) have gained rapid adoption in computational pathology for their ability to model long-range dependencies through self-attention, addressing the limitations of convolutional neural networks that excel at local pattern capture but struggle with global contextual reasoning. Recent pathology-specific foundation models have further advanced performance by leveraging large-scale pretraining. However, standard ViTs remain inherently non-equivariant to transformations such as rotations and reflections, which are ubiquitous variations in histopathology imaging. To address this limitation, we propose Equi-ViT, which integrates an equivariant convolution kernel into the patch embedding stage of a ViT architecture, imparting built-in rotational equivariance to learned representations. Equi-ViT achieves superior rotation-consistent patch embeddings and stable classification performance across image orientations. Our results on a public colorectal cancer dataset demonstrate that incorporating equivariant patch embedding enhances data efficiency and robustness, suggesting that equivariant transformers could potentially serve as more generalizable backbones for the application of ViT in histopathology, such as digital pathology foundation models.
CXR-LT 2024: A MICCAI challenge on long-tailed, multi-label, and zero-shot disease classification from chest X-ray
Jun 09, 2025Abstract:The CXR-LT series is a community-driven initiative designed to enhance lung disease classification using chest X-rays (CXR). It tackles challenges in open long-tailed lung disease classification and enhances the measurability of state-of-the-art techniques. The first event, CXR-LT 2023, aimed to achieve these goals by providing high-quality benchmark CXR data for model development and conducting comprehensive evaluations to identify ongoing issues impacting lung disease classification performance. Building on the success of CXR-LT 2023, the CXR-LT 2024 expands the dataset to 377,110 chest X-rays (CXRs) and 45 disease labels, including 19 new rare disease findings. It also introduces a new focus on zero-shot learning to address limitations identified in the previous event. Specifically, CXR-LT 2024 features three tasks: (i) long-tailed classification on a large, noisy test set, (ii) long-tailed classification on a manually annotated "gold standard" subset, and (iii) zero-shot generalization to five previously unseen disease findings. This paper provides an overview of CXR-LT 2024, detailing the data curation process and consolidating state-of-the-art solutions, including the use of multimodal models for rare disease detection, advanced generative approaches to handle noisy labels, and zero-shot learning strategies for unseen diseases. Additionally, the expanded dataset enhances disease coverage to better represent real-world clinical settings, offering a valuable resource for future research. By synthesizing the insights and innovations of participating teams, we aim to advance the development of clinically realistic and generalizable diagnostic models for chest radiography.
Equivariant Imaging Biomarkers for Robust Unsupervised Segmentation of Histopathology
May 08, 2025Abstract:Histopathology evaluation of tissue specimens through microscopic examination is essential for accurate disease diagnosis and prognosis. However, traditional manual analysis by specially trained pathologists is time-consuming, labor-intensive, cost-inefficient, and prone to inter-rater variability, potentially affecting diagnostic consistency and accuracy. As digital pathology images continue to proliferate, there is a pressing need for automated analysis to address these challenges. Recent advancements in artificial intelligence-based tools such as machine learning (ML) models, have significantly enhanced the precision and efficiency of analyzing histopathological slides. However, despite their impressive performance, ML models are invariant only to translation, lacking invariance to rotation and reflection. This limitation restricts their ability to generalize effectively, particularly in histopathology, where images intrinsically lack meaningful orientation. In this study, we develop robust, equivariant histopathological biomarkers through a novel symmetric convolutional kernel via unsupervised segmentation. The approach is validated using prostate tissue micro-array (TMA) images from 50 patients in the Gleason 2019 Challenge public dataset. The biomarkers extracted through this approach demonstrate enhanced robustness and generalizability against rotation compared to models using standard convolution kernels, holding promise for enhancing the accuracy, consistency, and robustness of ML models in digital pathology. Ultimately, this work aims to improve diagnostic and prognostic capabilities of histopathology beyond prostate cancer through equivariant imaging.
GMR-Conv: An Efficient Rotation and Reflection Equivariant Convolution Kernel Using Gaussian Mixture Rings
Apr 03, 2025Abstract:Symmetry, where certain features remain invariant under geometric transformations, can often serve as a powerful prior in designing convolutional neural networks (CNNs). While conventional CNNs inherently support translational equivariance, extending this property to rotation and reflection has proven challenging, often forcing a compromise between equivariance, efficiency, and information loss. In this work, we introduce Gaussian Mixture Ring Convolution (GMR-Conv), an efficient convolution kernel that smooths radial symmetry using a mixture of Gaussian-weighted rings. This design mitigates discretization errors of circular kernels, thereby preserving robust rotation and reflection equivariance without incurring computational overhead. We further optimize both the space and speed efficiency of GMR-Conv via a novel parameterization and computation strategy, allowing larger kernels at an acceptable cost. Extensive experiments on eight classification and one segmentation datasets demonstrate that GMR-Conv not only matches conventional CNNs' performance but can also surpass it in applications with orientation-less data. GMR-Conv is also proven to be more robust and efficient than the state-of-the-art equivariant learning methods. Our work provides inspiring empirical evidence that carefully applied radial symmetry can alleviate the challenges of information loss, marking a promising advance in equivariant network architectures. The code is available at https://github.com/XYPB/GMR-Conv.
Causal Modeling of fMRI Time-series for Interpretable Autism Spectrum Disorder Classification
Feb 21, 2025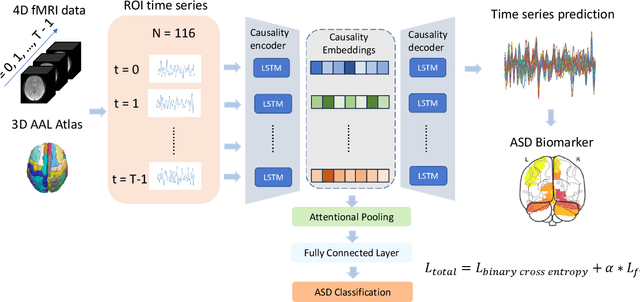
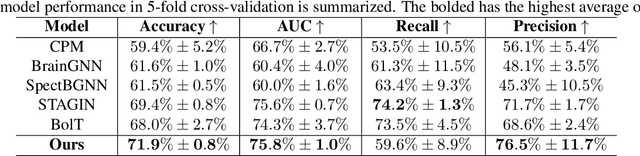
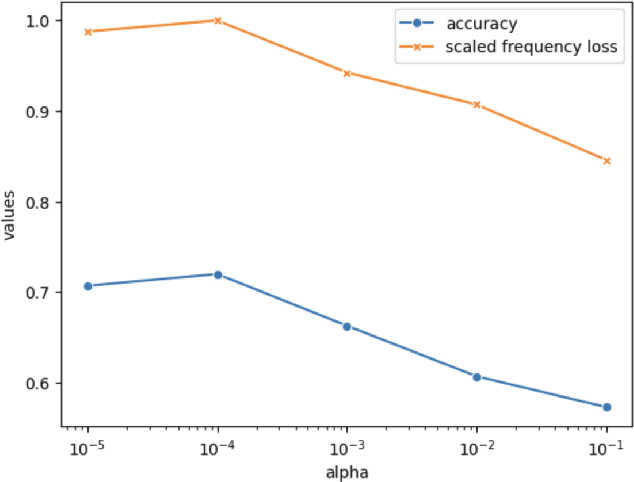
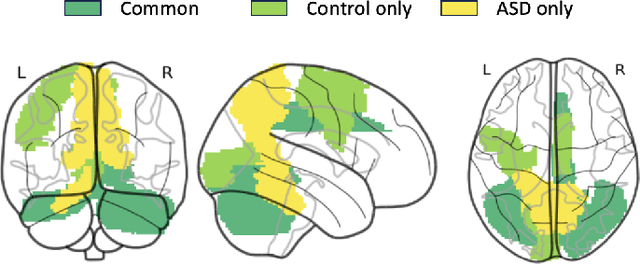
Abstract:Autism spectrum disorder (ASD) is a neurological and developmental disorder that affects social and communicative behaviors. It emerges in early life and is generally associated with lifelong disabilities. Thus, accurate and early diagnosis could facilitate treatment outcomes for those with ASD. Functional magnetic resonance imaging (fMRI) is a useful tool that measures changes in brain signaling to facilitate our understanding of ASD. Much effort is being made to identify ASD biomarkers using various connectome-based machine learning and deep learning classifiers. However, correlation-based models cannot capture the non-linear interactions between brain regions. To solve this problem, we introduce a causality-inspired deep learning model that uses time-series information from fMRI and captures causality among ROIs useful for ASD classification. The model is compared with other baseline and state-of-the-art models with 5-fold cross-validation on the ABIDE dataset. We filtered the dataset by choosing all the images with mean FD less than 15mm to ensure data quality. Our proposed model achieved the highest average classification accuracy of 71.9% and an average AUC of 75.8%. Moreover, the inter-ROI causality interpretation of the model suggests that the left precuneus, right precuneus, and cerebellum are placed in the top 10 ROIs in inter-ROI causality among the ASD population. In contrast, these ROIs are not ranked in the top 10 in the control population. We have validated our findings with the literature and found that abnormalities in these ROIs are often associated with ASD.
Improved Vessel Segmentation with Symmetric Rotation-Equivariant U-Net
Jan 24, 2025Abstract:Automated segmentation plays a pivotal role in medical image analysis and computer-assisted interventions. Despite the promising performance of existing methods based on convolutional neural networks (CNNs), they neglect useful equivariant properties for images, such as rotational and reflection equivariance. This limitation can decrease performance and lead to inconsistent predictions, especially in applications like vessel segmentation where explicit orientation is absent. While existing equivariant learning approaches attempt to mitigate these issues, they substantially increase learning cost, model size, or both. To overcome these challenges, we propose a novel application of an efficient symmetric rotation-equivariant (SRE) convolutional (SRE-Conv) kernel implementation to the U-Net architecture, to learn rotation and reflection-equivariant features, while also reducing the model size dramatically. We validate the effectiveness of our method through improved segmentation performance on retina vessel fundus imaging. Our proposed SRE U-Net not only significantly surpasses standard U-Net in handling rotated images, but also outperforms existing equivariant learning methods and does so with a reduced number of trainable parameters and smaller memory cost. The code is available at https://github.com/OnofreyLab/sre_conv_segm_isbi2025.
SRE-Conv: Symmetric Rotation Equivariant Convolution for Biomedical Image Classification
Jan 16, 2025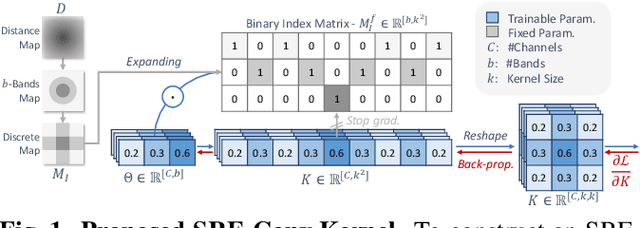
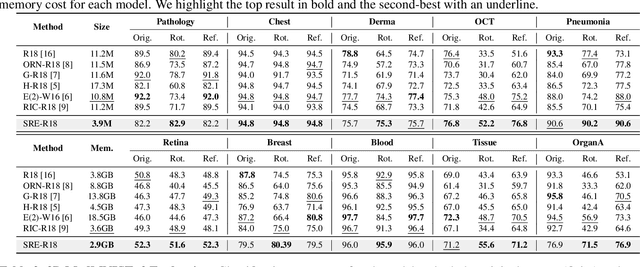
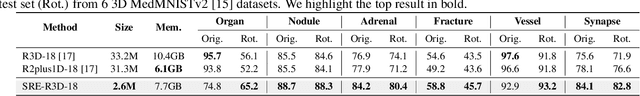
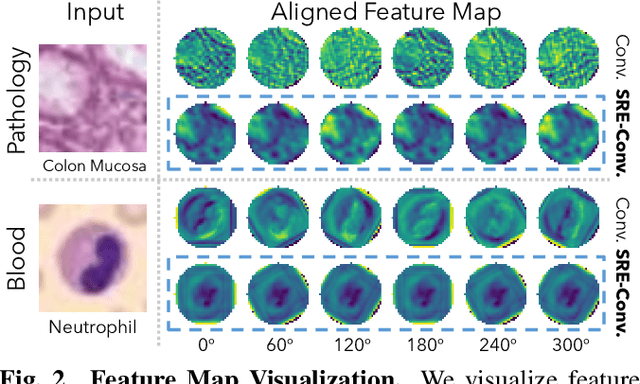
Abstract:Convolutional neural networks (CNNs) are essential tools for computer vision tasks, but they lack traditionally desired properties of extracted features that could further improve model performance, e.g., rotational equivariance. Such properties are ubiquitous in biomedical images, which often lack explicit orientation. While current work largely relies on data augmentation or explicit modules to capture orientation information, this comes at the expense of increased training costs or ineffective approximations of the desired equivariance. To overcome these challenges, we propose a novel and efficient implementation of the Symmetric Rotation-Equivariant (SRE) Convolution (SRE-Conv) kernel, designed to learn rotation-invariant features while simultaneously compressing the model size. The SRE-Conv kernel can easily be incorporated into any CNN backbone. We validate the ability of a deep SRE-CNN to capture equivariance to rotation using the public MedMNISTv2 dataset (16 total tasks). SRE-Conv-CNN demonstrated improved rotated image classification performance accuracy on all 16 test datasets in both 2D and 3D images, all while increasing efficiency with fewer parameters and reduced memory footprint. The code is available at https://github.com/XYPB/SRE-Conv.
Multi-View and Multi-Scale Alignment for Contrastive Language-Image Pre-training in Mammography
Sep 26, 2024



Abstract:Contrastive Language-Image Pre-training (CLIP) shows promise in medical image analysis but requires substantial data and computational resources. Due to these restrictions, existing CLIP applications in medical imaging focus mainly on modalities like chest X-rays that have abundant image-report data available, leaving many other important modalities under-explored. Here, we propose the first adaptation of the full CLIP model to mammography, which presents significant challenges due to labeled data scarcity, high-resolution images with small regions of interest, and data imbalance. We first develop a specialized supervision framework for mammography that leverages its multi-view nature. Furthermore, we design a symmetric local alignment module to better focus on detailed features in high-resolution images. Lastly, we incorporate a parameter-efficient fine-tuning approach for large language models pre-trained with medical knowledge to address data limitations. Our multi-view and multi-scale alignment (MaMA) method outperforms state-of-the-art baselines for three different tasks on two large real-world mammography datasets, EMBED and RSNA-Mammo, with only 52% model size compared with the largest baseline.
CLEFT: Language-Image Contrastive Learning with Efficient Large Language Model and Prompt Fine-Tuning
Jul 30, 2024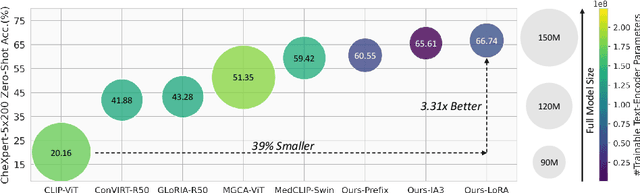
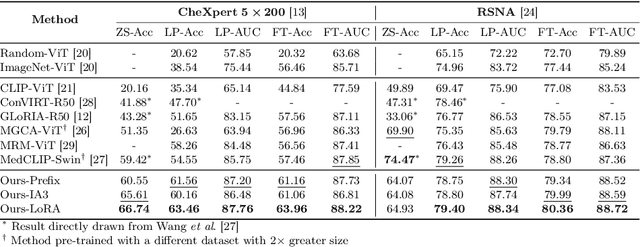
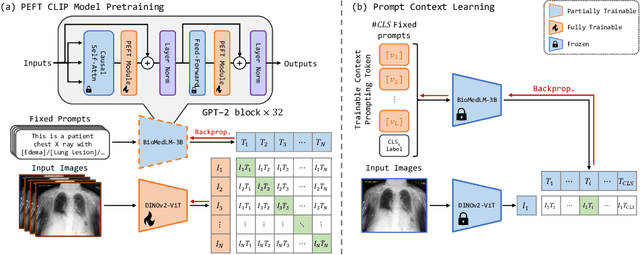
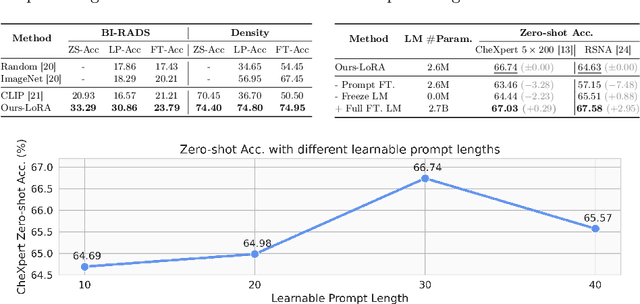
Abstract:Recent advancements in Contrastive Language-Image Pre-training (CLIP) have demonstrated notable success in self-supervised representation learning across various tasks. However, the existing CLIP-like approaches often demand extensive GPU resources and prolonged training times due to the considerable size of the model and dataset, making them poor for medical applications, in which large datasets are not always common. Meanwhile, the language model prompts are mainly manually derived from labels tied to images, potentially overlooking the richness of information within training samples. We introduce a novel language-image Contrastive Learning method with an Efficient large language model and prompt Fine-Tuning (CLEFT) that harnesses the strengths of the extensive pre-trained language and visual models. Furthermore, we present an efficient strategy for learning context-based prompts that mitigates the gap between informative clinical diagnostic data and simple class labels. Our method demonstrates state-of-the-art performance on multiple chest X-ray and mammography datasets compared with various baselines. The proposed parameter efficient framework can reduce the total trainable model size by 39% and reduce the trainable language model to only 4% compared with the current BERT encoder.
Assessment of Clonal Hematopoiesis of Indeterminate Potential from Cardiac Magnetic Resonance Imaging using Deep Learning in a Cardio-oncology Population
Jun 26, 2024Abstract:Background: We propose a novel method to identify who may likely have clonal hematopoiesis of indeterminate potential (CHIP), a condition characterized by the presence of somatic mutations in hematopoietic stem cells without detectable hematologic malignancy, using deep learning techniques. Methods: We developed a convolutional neural network (CNN) to predict CHIP status using 4 different views from standard delayed gadolinium-enhanced cardiac magnetic resonance imaging (CMR). We used 5-fold cross validation on 82 cardio-oncology patients to assess the performance of our model. Different algorithms were compared to find the optimal patient-level prediction method using the image-level CNN predictions. Results: We found that the best model had an area under the receiver operating characteristic curve of 0.85 and an accuracy of 82%. Conclusions: We conclude that a deep learning-based diagnostic approach for CHIP using CMR is promising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge