Tal Zeevi
Equivariant Imaging Biomarkers for Robust Unsupervised Segmentation of Histopathology
May 08, 2025Abstract:Histopathology evaluation of tissue specimens through microscopic examination is essential for accurate disease diagnosis and prognosis. However, traditional manual analysis by specially trained pathologists is time-consuming, labor-intensive, cost-inefficient, and prone to inter-rater variability, potentially affecting diagnostic consistency and accuracy. As digital pathology images continue to proliferate, there is a pressing need for automated analysis to address these challenges. Recent advancements in artificial intelligence-based tools such as machine learning (ML) models, have significantly enhanced the precision and efficiency of analyzing histopathological slides. However, despite their impressive performance, ML models are invariant only to translation, lacking invariance to rotation and reflection. This limitation restricts their ability to generalize effectively, particularly in histopathology, where images intrinsically lack meaningful orientation. In this study, we develop robust, equivariant histopathological biomarkers through a novel symmetric convolutional kernel via unsupervised segmentation. The approach is validated using prostate tissue micro-array (TMA) images from 50 patients in the Gleason 2019 Challenge public dataset. The biomarkers extracted through this approach demonstrate enhanced robustness and generalizability against rotation compared to models using standard convolution kernels, holding promise for enhancing the accuracy, consistency, and robustness of ML models in digital pathology. Ultimately, this work aims to improve diagnostic and prognostic capabilities of histopathology beyond prostate cancer through equivariant imaging.
Enhancing Uncertainty Estimation in Semantic Segmentation via Monte-Carlo Frequency Dropout
Jan 20, 2025



Abstract:Monte-Carlo (MC) Dropout provides a practical solution for estimating predictive distributions in deterministic neural networks. Traditional dropout, applied within the signal space, may fail to account for frequency-related noise common in medical imaging, leading to biased predictive estimates. A novel approach extends Dropout to the frequency domain, allowing stochastic attenuation of signal frequencies during inference. This creates diverse global textural variations in feature maps while preserving structural integrity -- a factor we hypothesize and empirically show is contributing to accurately estimating uncertainties in semantic segmentation. We evaluated traditional MC-Dropout and the MC-frequency Dropout in three segmentation tasks involving different imaging modalities: (i) prostate zones in biparametric MRI, (ii) liver tumors in contrast-enhanced CT, and (iii) lungs in chest X-ray scans. Our results show that MC-Frequency Dropout improves calibration, convergence, and semantic uncertainty, thereby improving prediction scrutiny, boundary delineation, and has the potential to enhance medical decision-making.
SRE-Conv: Symmetric Rotation Equivariant Convolution for Biomedical Image Classification
Jan 16, 2025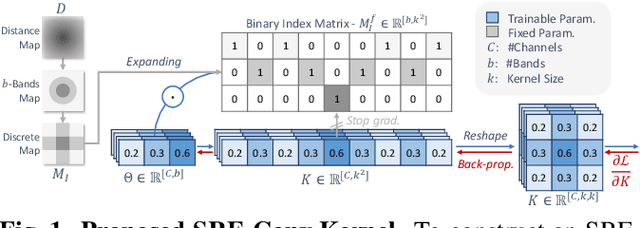
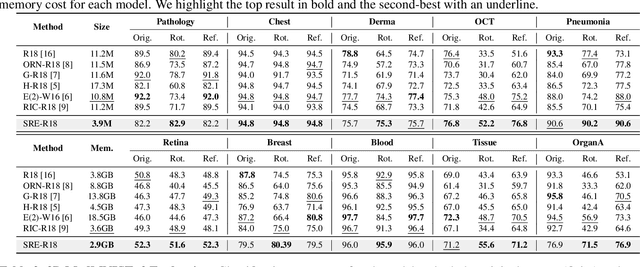
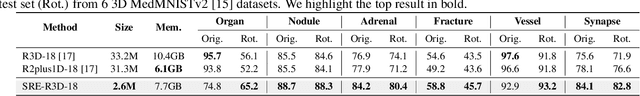
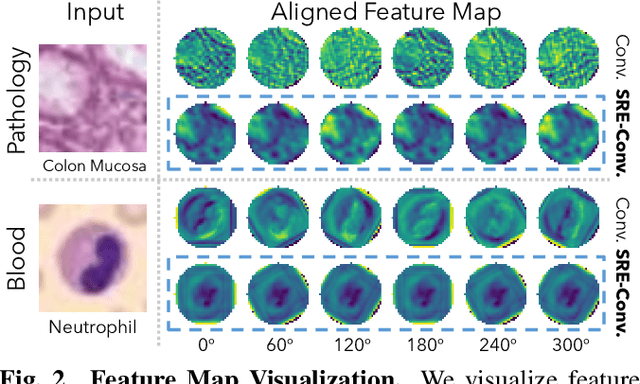
Abstract:Convolutional neural networks (CNNs) are essential tools for computer vision tasks, but they lack traditionally desired properties of extracted features that could further improve model performance, e.g., rotational equivariance. Such properties are ubiquitous in biomedical images, which often lack explicit orientation. While current work largely relies on data augmentation or explicit modules to capture orientation information, this comes at the expense of increased training costs or ineffective approximations of the desired equivariance. To overcome these challenges, we propose a novel and efficient implementation of the Symmetric Rotation-Equivariant (SRE) Convolution (SRE-Conv) kernel, designed to learn rotation-invariant features while simultaneously compressing the model size. The SRE-Conv kernel can easily be incorporated into any CNN backbone. We validate the ability of a deep SRE-CNN to capture equivariance to rotation using the public MedMNISTv2 dataset (16 total tasks). SRE-Conv-CNN demonstrated improved rotated image classification performance accuracy on all 16 test datasets in both 2D and 3D images, all while increasing efficiency with fewer parameters and reduced memory footprint. The code is available at https://github.com/XYPB/SRE-Conv.
Rate-In: Information-Driven Adaptive Dropout Rates for Improved Inference-Time Uncertainty Estimation
Dec 10, 2024



Abstract:Accurate uncertainty estimation is crucial for deploying neural networks in risk-sensitive applications such as medical diagnosis. Monte Carlo Dropout is a widely used technique for approximating predictive uncertainty by performing stochastic forward passes with dropout during inference. However, using static dropout rates across all layers and inputs can lead to suboptimal uncertainty estimates, as it fails to adapt to the varying characteristics of individual inputs and network layers. Existing approaches optimize dropout rates during training using labeled data, resulting in fixed inference-time parameters that cannot adjust to new data distributions, compromising uncertainty estimates in Monte Carlo simulations. In this paper, we propose Rate-In, an algorithm that dynamically adjusts dropout rates during inference by quantifying the information loss induced by dropout in each layer's feature maps. By treating dropout as controlled noise injection and leveraging information-theoretic principles, Rate-In adapts dropout rates per layer and per input instance without requiring ground truth labels. By quantifying the functional information loss in feature maps, we adaptively tune dropout rates to maintain perceptual quality across diverse medical imaging tasks and architectural configurations. Our extensive empirical study on synthetic data and real-world medical imaging tasks demonstrates that Rate-In improves calibration and sharpens uncertainty estimates compared to fixed or heuristic dropout rates without compromising predictive performance. Rate-In offers a practical, unsupervised, inference-time approach to optimizing dropout for more reliable predictive uncertainty estimation in critical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge