Jiazhen Zhang
PET Head Motion Estimation Using Supervised Deep Learning with Attention
Oct 14, 2025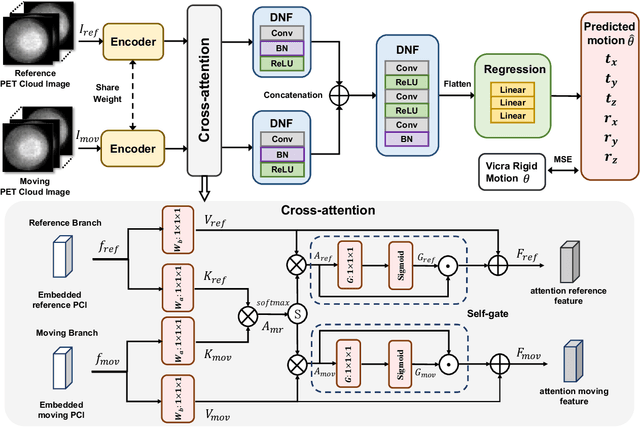
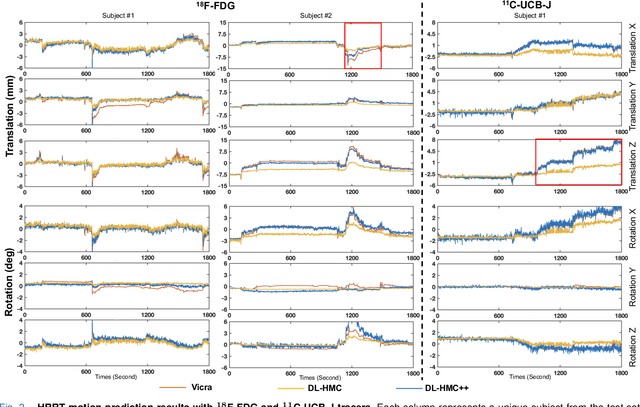
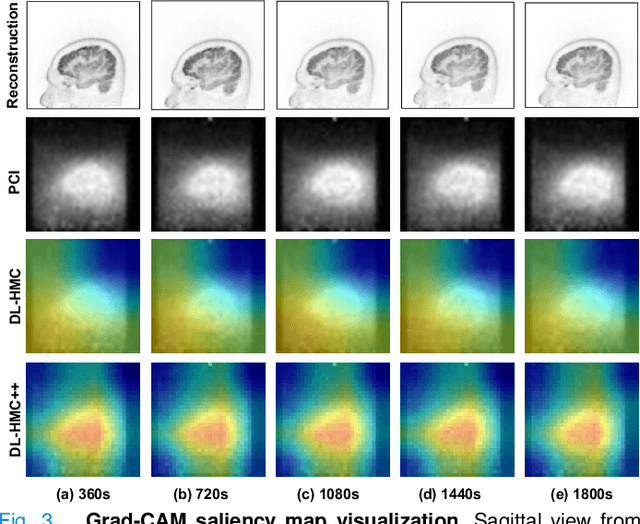
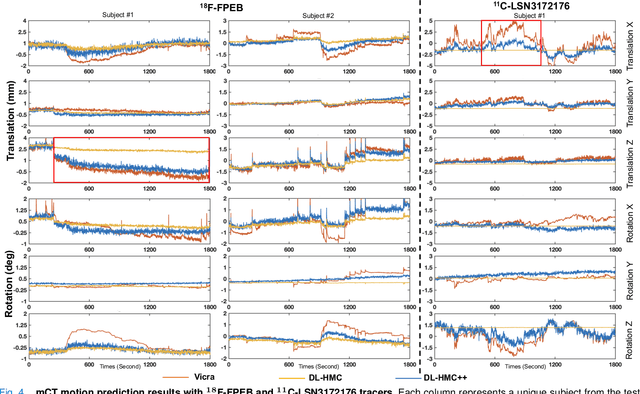
Abstract:Head movement poses a significant challenge in brain positron emission tomography (PET) imaging, resulting in image artifacts and tracer uptake quantification inaccuracies. Effective head motion estimation and correction are crucial for precise quantitative image analysis and accurate diagnosis of neurological disorders. Hardware-based motion tracking (HMT) has limited applicability in real-world clinical practice. To overcome this limitation, we propose a deep-learning head motion correction approach with cross-attention (DL-HMC++) to predict rigid head motion from one-second 3D PET raw data. DL-HMC++ is trained in a supervised manner by leveraging existing dynamic PET scans with gold-standard motion measurements from external HMT. We evaluate DL-HMC++ on two PET scanners (HRRT and mCT) and four radiotracers (18F-FDG, 18F-FPEB, 11C-UCB-J, and 11C-LSN3172176) to demonstrate the effectiveness and generalization of the approach in large cohort PET studies. Quantitative and qualitative results demonstrate that DL-HMC++ consistently outperforms state-of-the-art data-driven motion estimation methods, producing motion-free images with clear delineation of brain structures and reduced motion artifacts that are indistinguishable from gold-standard HMT. Brain region of interest standard uptake value analysis exhibits average difference ratios between DL-HMC++ and gold-standard HMT to be 1.2 plus-minus 0.5% for HRRT and 0.5 plus-minus 0.2% for mCT. DL-HMC++ demonstrates the potential for data-driven PET head motion correction to remove the burden of HMT, making motion correction accessible to clinical populations beyond research settings. The code is available at https://github.com/maxxxxxxcai/DL-HMC-TMI.
GMR-Conv: An Efficient Rotation and Reflection Equivariant Convolution Kernel Using Gaussian Mixture Rings
Apr 03, 2025Abstract:Symmetry, where certain features remain invariant under geometric transformations, can often serve as a powerful prior in designing convolutional neural networks (CNNs). While conventional CNNs inherently support translational equivariance, extending this property to rotation and reflection has proven challenging, often forcing a compromise between equivariance, efficiency, and information loss. In this work, we introduce Gaussian Mixture Ring Convolution (GMR-Conv), an efficient convolution kernel that smooths radial symmetry using a mixture of Gaussian-weighted rings. This design mitigates discretization errors of circular kernels, thereby preserving robust rotation and reflection equivariance without incurring computational overhead. We further optimize both the space and speed efficiency of GMR-Conv via a novel parameterization and computation strategy, allowing larger kernels at an acceptable cost. Extensive experiments on eight classification and one segmentation datasets demonstrate that GMR-Conv not only matches conventional CNNs' performance but can also surpass it in applications with orientation-less data. GMR-Conv is also proven to be more robust and efficient than the state-of-the-art equivariant learning methods. Our work provides inspiring empirical evidence that carefully applied radial symmetry can alleviate the challenges of information loss, marking a promising advance in equivariant network architectures. The code is available at https://github.com/XYPB/GMR-Conv.
Improved Vessel Segmentation with Symmetric Rotation-Equivariant U-Net
Jan 24, 2025Abstract:Automated segmentation plays a pivotal role in medical image analysis and computer-assisted interventions. Despite the promising performance of existing methods based on convolutional neural networks (CNNs), they neglect useful equivariant properties for images, such as rotational and reflection equivariance. This limitation can decrease performance and lead to inconsistent predictions, especially in applications like vessel segmentation where explicit orientation is absent. While existing equivariant learning approaches attempt to mitigate these issues, they substantially increase learning cost, model size, or both. To overcome these challenges, we propose a novel application of an efficient symmetric rotation-equivariant (SRE) convolutional (SRE-Conv) kernel implementation to the U-Net architecture, to learn rotation and reflection-equivariant features, while also reducing the model size dramatically. We validate the effectiveness of our method through improved segmentation performance on retina vessel fundus imaging. Our proposed SRE U-Net not only significantly surpasses standard U-Net in handling rotated images, but also outperforms existing equivariant learning methods and does so with a reduced number of trainable parameters and smaller memory cost. The code is available at https://github.com/OnofreyLab/sre_conv_segm_isbi2025.
SRE-Conv: Symmetric Rotation Equivariant Convolution for Biomedical Image Classification
Jan 16, 2025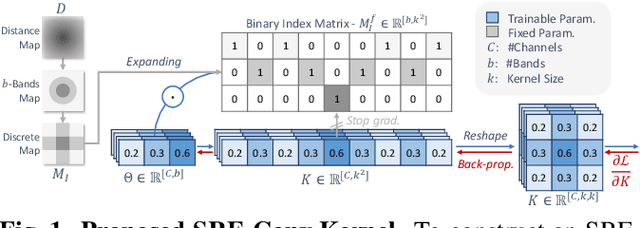
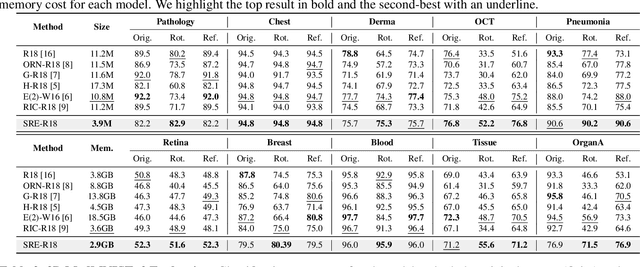
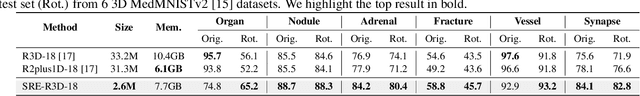
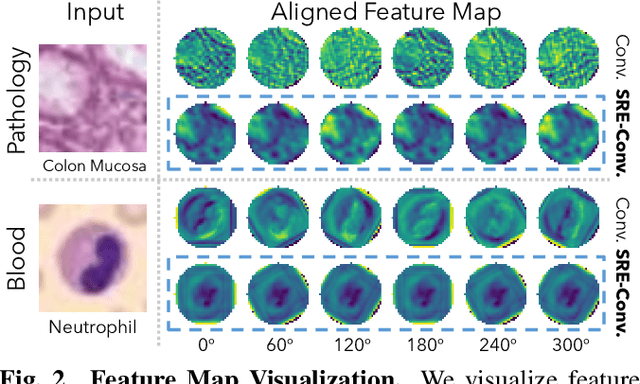
Abstract:Convolutional neural networks (CNNs) are essential tools for computer vision tasks, but they lack traditionally desired properties of extracted features that could further improve model performance, e.g., rotational equivariance. Such properties are ubiquitous in biomedical images, which often lack explicit orientation. While current work largely relies on data augmentation or explicit modules to capture orientation information, this comes at the expense of increased training costs or ineffective approximations of the desired equivariance. To overcome these challenges, we propose a novel and efficient implementation of the Symmetric Rotation-Equivariant (SRE) Convolution (SRE-Conv) kernel, designed to learn rotation-invariant features while simultaneously compressing the model size. The SRE-Conv kernel can easily be incorporated into any CNN backbone. We validate the ability of a deep SRE-CNN to capture equivariance to rotation using the public MedMNISTv2 dataset (16 total tasks). SRE-Conv-CNN demonstrated improved rotated image classification performance accuracy on all 16 test datasets in both 2D and 3D images, all while increasing efficiency with fewer parameters and reduced memory footprint. The code is available at https://github.com/XYPB/SRE-Conv.
Head Motion Degrades Machine Learning Classification of Alzheimer's Disease from Positron Emission Tomography
Jan 14, 2025



Abstract:Brain positron emission tomography (PET) imaging is broadly used in research and clinical routines to study, diagnose, and stage Alzheimer's disease (AD). However, its potential cannot be fully exploited yet due to the lack of portable motion correction solutions, especially in clinical settings. Head motion during data acquisition has indeed been shown to degrade image quality and induces tracer uptake quantification error. In this study, we demonstrate that it also biases machine learning-based AD classification. We start by proposing a binary classification algorithm solely based on PET images. We find that it reaches a high accuracy in classifying motion corrected images into cognitive normal or AD. We demonstrate that the classification accuracy substantially decreases when images lack motion correction, thereby limiting the algorithm's effectiveness and biasing image interpretation. We validate these findings in cohorts of 128 $^{11}$C-UCB-J and 173 $^{18}$F-FDG scans, two tracers highly relevant to the study of AD. Classification accuracies decreased by 10% and 5% on 20 $^{18}$F-FDG and 20 $^{11}$C-UCB-J testing cases, respectively. Our findings underscore the critical need for efficient motion correction methods to make the most of the diagnostic capabilities of PET-based machine learning.
Fast-MC-PET: A Novel Deep Learning-aided Motion Correction and Reconstruction Framework for Accelerated PET
Feb 14, 2023



Abstract:Patient motion during PET is inevitable. Its long acquisition time not only increases the motion and the associated artifacts but also the patient's discomfort, thus PET acceleration is desirable. However, accelerating PET acquisition will result in reconstructed images with low SNR, and the image quality will still be degraded by motion-induced artifacts. Most of the previous PET motion correction methods are motion type specific that require motion modeling, thus may fail when multiple types of motion present together. Also, those methods are customized for standard long acquisition and could not be directly applied to accelerated PET. To this end, modeling-free universal motion correction reconstruction for accelerated PET is still highly under-explored. In this work, we propose a novel deep learning-aided motion correction and reconstruction framework for accelerated PET, called Fast-MC-PET. Our framework consists of a universal motion correction (UMC) and a short-to-long acquisition reconstruction (SL-Reon) module. The UMC enables modeling-free motion correction by estimating quasi-continuous motion from ultra-short frame reconstructions and using this information for motion-compensated reconstruction. Then, the SL-Recon converts the accelerated UMC image with low counts to a high-quality image with high counts for our final reconstruction output. Our experimental results on human studies show that our Fast-MC-PET can enable 7-fold acceleration and use only 2 minutes acquisition to generate high-quality reconstruction images that outperform/match previous motion correction reconstruction methods using standard 15 minutes long acquisition data.
Dual-Branch Squeeze-Fusion-Excitation Module for Cross-Modality Registration of Cardiac SPECT and CT
Jun 10, 2022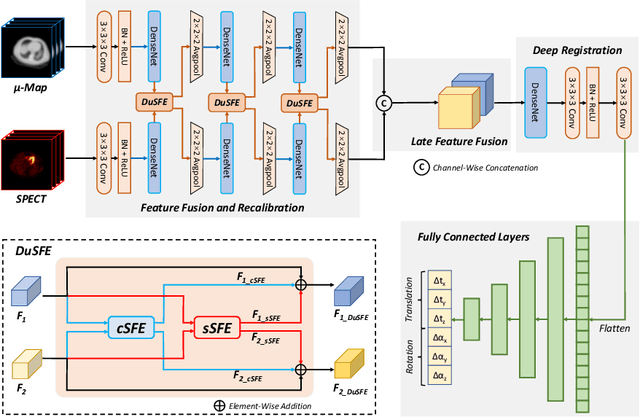
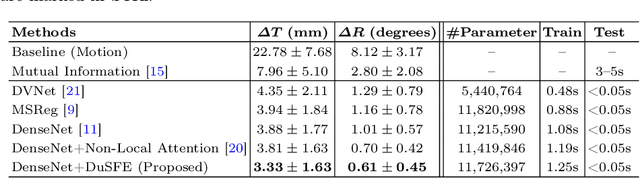
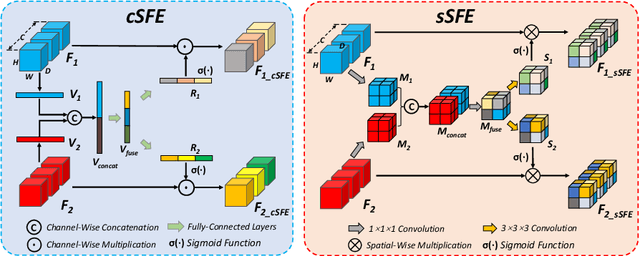
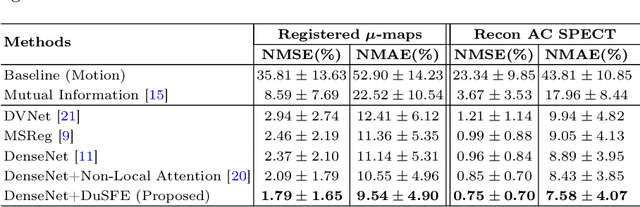
Abstract:Single-photon emission computed tomography (SPECT) is a widely applied imaging approach for diagnosis of coronary artery diseases. Attenuation maps (u-maps) derived from computed tomography (CT) are utilized for attenuation correction (AC) to improve diagnostic accuracy of cardiac SPECT. However, SPECT and CT are obtained sequentially in clinical practice, which potentially induces misregistration between the two scans. Convolutional neural networks (CNN) are powerful tools for medical image registration. Previous CNN-based methods for cross-modality registration either directly concatenated two input modalities as an early feature fusion or extracted image features using two separate CNN modules for a late fusion. These methods do not fully extract or fuse the cross-modality information. Besides, deep-learning-based rigid registration of cardiac SPECT and CT-derived u-maps has not been investigated before. In this paper, we propose a Dual-Branch Squeeze-Fusion-Excitation (DuSFE) module for the registration of cardiac SPECT and CT-derived u-maps. DuSFE fuses the knowledge from multiple modalities to recalibrate both channel-wise and spatial features for each modality. DuSFE can be embedded at multiple convolutional layers to enable feature fusion at different spatial dimensions. Our studies using clinical data demonstrated that a network embedded with DuSFE generated substantial lower registration errors and therefore more accurate AC SPECT images than previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge