Xiongchao Chen
A Generalizable 3D Diffusion Framework for Low-Dose and Few-View Cardiac SPECT
Dec 21, 2024Abstract:Myocardial perfusion imaging using SPECT is widely utilized to diagnose coronary artery diseases, but image quality can be negatively affected in low-dose and few-view acquisition settings. Although various deep learning methods have been introduced to improve image quality from low-dose or few-view SPECT data, previous approaches often fail to generalize across different acquisition settings, limiting their applicability in reality. This work introduced DiffSPECT-3D, a diffusion framework for 3D cardiac SPECT imaging that effectively adapts to different acquisition settings without requiring further network re-training or fine-tuning. Using both image and projection data, a consistency strategy is proposed to ensure that diffusion sampling at each step aligns with the low-dose/few-view projection measurements, the image data, and the scanner geometry, thus enabling generalization to different low-dose/few-view settings. Incorporating anatomical spatial information from CT and total variation constraint, we proposed a 2.5D conditional strategy to allow the DiffSPECT-3D to observe 3D contextual information from the entire image volume, addressing the 3D memory issues in diffusion model. We extensively evaluated the proposed method on 1,325 clinical 99mTc tetrofosmin stress/rest studies from 795 patients. Each study was reconstructed into 5 different low-count and 5 different few-view levels for model evaluations, ranging from 1% to 50% and from 1 view to 9 view, respectively. Validated against cardiac catheterization results and diagnostic comments from nuclear cardiologists, the presented results show the potential to achieve low-dose and few-view SPECT imaging without compromising clinical performance. Additionally, DiffSPECT-3D could be directly applied to full-dose SPECT images to further improve image quality, especially in a low-dose stress-first cardiac SPECT imaging protocol.
Noise-aware Dynamic Image Denoising and Positron Range Correction for Rubidium-82 Cardiac PET Imaging via Self-supervision
Sep 17, 2024



Abstract:Rb-82 is a radioactive isotope widely used for cardiac PET imaging. Despite numerous benefits of 82-Rb, there are several factors that limits its image quality and quantitative accuracy. First, the short half-life of 82-Rb results in noisy dynamic frames. Low signal-to-noise ratio would result in inaccurate and biased image quantification. Noisy dynamic frames also lead to highly noisy parametric images. The noise levels also vary substantially in different dynamic frames due to radiotracer decay and short half-life. Existing denoising methods are not applicable for this task due to the lack of paired training inputs/labels and inability to generalize across varying noise levels. Second, 82-Rb emits high-energy positrons. Compared with other tracers such as 18-F, 82-Rb travels a longer distance before annihilation, which negatively affect image spatial resolution. Here, the goal of this study is to propose a self-supervised method for simultaneous (1) noise-aware dynamic image denoising and (2) positron range correction for 82-Rb cardiac PET imaging. Tested on a series of PET scans from a cohort of normal volunteers, the proposed method produced images with superior visual quality. To demonstrate the improvement in image quantification, we compared image-derived input functions (IDIFs) with arterial input functions (AIFs) from continuous arterial blood samples. The IDIF derived from the proposed method led to lower AUC differences, decreasing from 11.09% to 7.58% on average, compared to the original dynamic frames. The proposed method also improved the quantification of myocardium blood flow (MBF), as validated against 15-O-water scans, with mean MBF differences decreased from 0.43 to 0.09, compared to the original dynamic frames. We also conducted a generalizability experiment on 37 patient scans obtained from a different country using a different scanner.
2.5D Multi-view Averaging Diffusion Model for 3D Medical Image Translation: Application to Low-count PET Reconstruction with CT-less Attenuation Correction
Jun 12, 2024
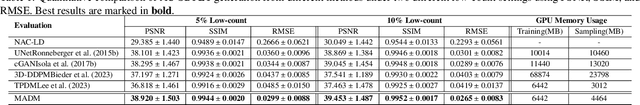

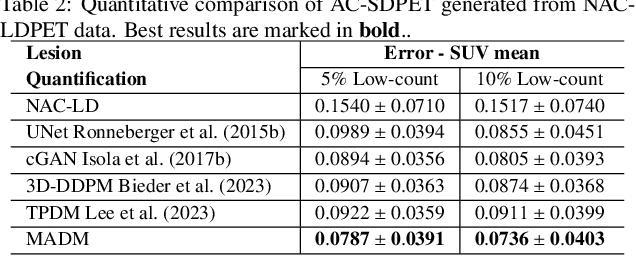
Abstract:Positron Emission Tomography (PET) is an important clinical imaging tool but inevitably introduces radiation hazards to patients and healthcare providers. Reducing the tracer injection dose and eliminating the CT acquisition for attenuation correction can reduce the overall radiation dose, but often results in PET with high noise and bias. Thus, it is desirable to develop 3D methods to translate the non-attenuation-corrected low-dose PET (NAC-LDPET) into attenuation-corrected standard-dose PET (AC-SDPET). Recently, diffusion models have emerged as a new state-of-the-art deep learning method for image-to-image translation, better than traditional CNN-based methods. However, due to the high computation cost and memory burden, it is largely limited to 2D applications. To address these challenges, we developed a novel 2.5D Multi-view Averaging Diffusion Model (MADM) for 3D image-to-image translation with application on NAC-LDPET to AC-SDPET translation. Specifically, MADM employs separate diffusion models for axial, coronal, and sagittal views, whose outputs are averaged in each sampling step to ensure the 3D generation quality from multiple views. To accelerate the 3D sampling process, we also proposed a strategy to use the CNN-based 3D generation as a prior for the diffusion model. Our experimental results on human patient studies suggested that MADM can generate high-quality 3D translation images, outperforming previous CNN-based and Diffusion-based baseline methods.
TAI-GAN: A Temporally and Anatomically Informed Generative Adversarial Network for early-to-late frame conversion in dynamic cardiac PET inter-frame motion correction
Feb 14, 2024
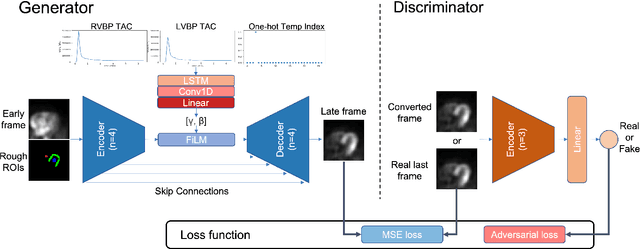
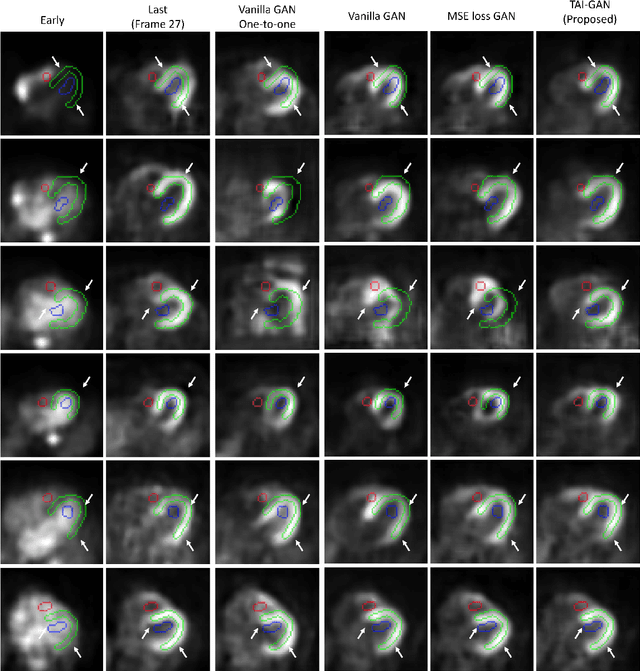
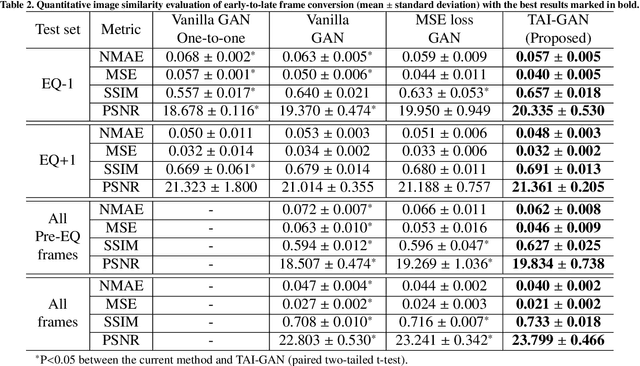
Abstract:Inter-frame motion in dynamic cardiac positron emission tomography (PET) using rubidium-82 (82-Rb) myocardial perfusion imaging impacts myocardial blood flow (MBF) quantification and the diagnosis accuracy of coronary artery diseases. However, the high cross-frame distribution variation due to rapid tracer kinetics poses a considerable challenge for inter-frame motion correction, especially for early frames where intensity-based image registration techniques often fail. To address this issue, we propose a novel method called Temporally and Anatomically Informed Generative Adversarial Network (TAI-GAN) that utilizes an all-to-one mapping to convert early frames into those with tracer distribution similar to the last reference frame. The TAI-GAN consists of a feature-wise linear modulation layer that encodes channel-wise parameters generated from temporal information and rough cardiac segmentation masks with local shifts that serve as anatomical information. Our proposed method was evaluated on a clinical 82-Rb PET dataset, and the results show that our TAI-GAN can produce converted early frames with high image quality, comparable to the real reference frames. After TAI-GAN conversion, the motion estimation accuracy and subsequent myocardial blood flow (MBF) quantification with both conventional and deep learning-based motion correction methods were improved compared to using the original frames.
POUR-Net: A Population-Prior-Aided Over-Under-Representation Network for Low-Count PET Attenuation Map Generation
Jan 25, 2024



Abstract:Low-dose PET offers a valuable means of minimizing radiation exposure in PET imaging. However, the prevalent practice of employing additional CT scans for generating attenuation maps (u-map) for PET attenuation correction significantly elevates radiation doses. To address this concern and further mitigate radiation exposure in low-dose PET exams, we propose POUR-Net - an innovative population-prior-aided over-under-representation network that aims for high-quality attenuation map generation from low-dose PET. First, POUR-Net incorporates an over-under-representation network (OUR-Net) to facilitate efficient feature extraction, encompassing both low-resolution abstracted and fine-detail features, for assisting deep generation on the full-resolution level. Second, complementing OUR-Net, a population prior generation machine (PPGM) utilizing a comprehensive CT-derived u-map dataset, provides additional prior information to aid OUR-Net generation. The integration of OUR-Net and PPGM within a cascade framework enables iterative refinement of $\mu$-map generation, resulting in the production of high-quality $\mu$-maps. Experimental results underscore the effectiveness of POUR-Net, showing it as a promising solution for accurate CT-free low-count PET attenuation correction, which also surpasses the performance of previous baseline methods.
Dual-Domain Coarse-to-Fine Progressive Estimation Network for Simultaneous Denoising, Limited-View Reconstruction, and Attenuation Correction of Cardiac SPECT
Jan 23, 2024Abstract:Single-Photon Emission Computed Tomography (SPECT) is widely applied for the diagnosis of coronary artery diseases. Low-dose (LD) SPECT aims to minimize radiation exposure but leads to increased image noise. Limited-view (LV) SPECT, such as the latest GE MyoSPECT ES system, enables accelerated scanning and reduces hardware expenses but degrades reconstruction accuracy. Additionally, Computed Tomography (CT) is commonly used to derive attenuation maps ($\mu$-maps) for attenuation correction (AC) of cardiac SPECT, but it will introduce additional radiation exposure and SPECT-CT misalignments. Although various methods have been developed to solely focus on LD denoising, LV reconstruction, or CT-free AC in SPECT, the solution for simultaneously addressing these tasks remains challenging and under-explored. Furthermore, it is essential to explore the potential of fusing cross-domain and cross-modality information across these interrelated tasks to further enhance the accuracy of each task. Thus, we propose a Dual-Domain Coarse-to-Fine Progressive Network (DuDoCFNet), a multi-task learning method for simultaneous LD denoising, LV reconstruction, and CT-free $\mu$-map generation of cardiac SPECT. Paired dual-domain networks in DuDoCFNet are cascaded using a multi-layer fusion mechanism for cross-domain and cross-modality feature fusion. Two-stage progressive learning strategies are applied in both projection and image domains to achieve coarse-to-fine estimations of SPECT projections and CT-derived $\mu$-maps. Our experiments demonstrate DuDoCFNet's superior accuracy in estimating projections, generating $\mu$-maps, and AC reconstructions compared to existing single- or multi-task learning methods, under various iterations and LD levels. The source code of this work is available at https://github.com/XiongchaoChen/DuDoCFNet-MultiTask.
Dose-aware Diffusion Model for 3D Ultra Low-dose PET Imaging
Nov 07, 2023Abstract:As PET imaging is accompanied by substantial radiation exposure and cancer risk, reducing radiation dose in PET scans is an important topic. Recently, diffusion models have emerged as the new state-of-the-art generative model to generate high-quality samples and have demonstrated strong potential for various tasks in medical imaging. However, it is difficult to extend diffusion models for 3D image reconstructions due to the memory burden. Directly stacking 2D slices together to create 3D image volumes would results in severe inconsistencies between slices. Previous works tried to either applying a penalty term along the z-axis to remove inconsistencies or reconstructing the 3D image volumes with 2 pre-trained perpendicular 2D diffusion models. Nonetheless, these previous methods failed to produce satisfactory results in challenging cases for PET image denoising. In addition to administered dose, the noise-levels in PET images are affected by several other factors in clinical settings, such as scan time, patient size, and weight, etc. Therefore, a method to simultaneously denoise PET images with different noise-levels is needed. Here, we proposed a dose-aware diffusion model for 3D low-dose PET imaging (DDPET) to address these challenges. The proposed DDPET method was tested on 295 patients from three different medical institutions globally with different low-dose levels. These patient data were acquired on three different commercial PET scanners, including Siemens Vision Quadra, Siemens mCT, and United Imaging Healthcare uExplorere. The proposed method demonstrated superior performance over previously proposed diffusion models for 3D imaging problems as well as models proposed for noise-aware medical image denoising. Code is available at: xxx.
TAI-GAN: Temporally and Anatomically Informed GAN for early-to-late frame conversion in dynamic cardiac PET motion correction
Aug 23, 2023Abstract:The rapid tracer kinetics of rubidium-82 ($^{82}$Rb) and high variation of cross-frame distribution in dynamic cardiac positron emission tomography (PET) raise significant challenges for inter-frame motion correction, particularly for the early frames where conventional intensity-based image registration techniques are not applicable. Alternatively, a promising approach utilizes generative methods to handle the tracer distribution changes to assist existing registration methods. To improve frame-wise registration and parametric quantification, we propose a Temporally and Anatomically Informed Generative Adversarial Network (TAI-GAN) to transform the early frames into the late reference frame using an all-to-one mapping. Specifically, a feature-wise linear modulation layer encodes channel-wise parameters generated from temporal tracer kinetics information, and rough cardiac segmentations with local shifts serve as the anatomical information. We validated our proposed method on a clinical $^{82}$Rb PET dataset and found that our TAI-GAN can produce converted early frames with high image quality, comparable to the real reference frames. After TAI-GAN conversion, motion estimation accuracy and clinical myocardial blood flow (MBF) quantification were improved compared to using the original frames. Our code is published at https://github.com/gxq1998/TAI-GAN.
Transformer-based Dual-domain Network for Few-view Dedicated Cardiac SPECT Image Reconstructions
Jul 23, 2023
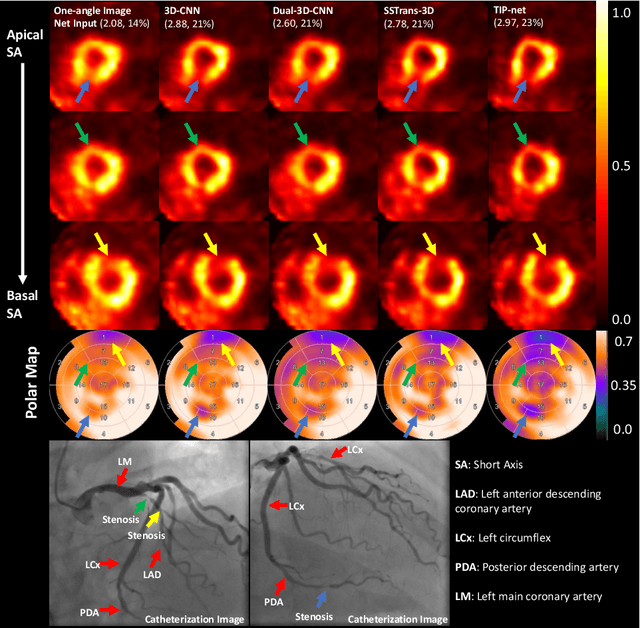
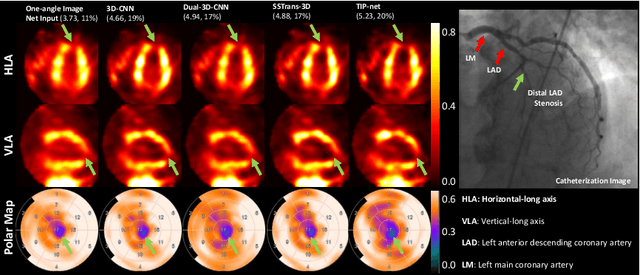
Abstract:Cardiovascular disease (CVD) is the leading cause of death worldwide, and myocardial perfusion imaging using SPECT has been widely used in the diagnosis of CVDs. The GE 530/570c dedicated cardiac SPECT scanners adopt a stationary geometry to simultaneously acquire 19 projections to increase sensitivity and achieve dynamic imaging. However, the limited amount of angular sampling negatively affects image quality. Deep learning methods can be implemented to produce higher-quality images from stationary data. This is essentially a few-view imaging problem. In this work, we propose a novel 3D transformer-based dual-domain network, called TIP-Net, for high-quality 3D cardiac SPECT image reconstructions. Our method aims to first reconstruct 3D cardiac SPECT images directly from projection data without the iterative reconstruction process by proposing a customized projection-to-image domain transformer. Then, given its reconstruction output and the original few-view reconstruction, we further refine the reconstruction using an image-domain reconstruction network. Validated by cardiac catheterization images, diagnostic interpretations from nuclear cardiologists, and defect size quantified by an FDA 510(k)-cleared clinical software, our method produced images with higher cardiac defect contrast on human studies compared with previous baseline methods, potentially enabling high-quality defect visualization using stationary few-view dedicated cardiac SPECT scanners.
Cross-domain Iterative Network for Simultaneous Denoising, Limited-angle Reconstruction, and Attenuation Correction of Low-dose Cardiac SPECT
May 17, 2023



Abstract:Single-Photon Emission Computed Tomography (SPECT) is widely applied for the diagnosis of ischemic heart diseases. Low-dose (LD) SPECT aims to minimize radiation exposure but leads to increased image noise. Limited-angle (LA) SPECT enables faster scanning and reduced hardware costs but results in lower reconstruction accuracy. Additionally, computed tomography (CT)-derived attenuation maps ($\mu$-maps) are commonly used for SPECT attenuation correction (AC), but it will cause extra radiation exposure and SPECT-CT misalignments. In addition, the majority of SPECT scanners in the market are not hybrid SPECT/CT scanners. Although various deep learning methods have been introduced to separately address these limitations, the solution for simultaneously addressing these challenges still remains highly under-explored and challenging. To this end, we propose a Cross-domain Iterative Network (CDI-Net) for simultaneous denoising, LA reconstruction, and CT-free AC in cardiac SPECT. In CDI-Net, paired projection- and image-domain networks are end-to-end connected to fuse the emission and anatomical information across domains and iterations. Adaptive Weight Recalibrators (AWR) adjust the multi-channel input features to enhance prediction accuracy. Our experiments using clinical data showed that CDI-Net produced more accurate $\mu$-maps, projections, and reconstructions compared to existing approaches that addressed each task separately. Ablation studies demonstrated the significance of cross-domain and cross-iteration connections, as well as AWR, in improving the reconstruction performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge