Menghua Xia
Fed-NDIF: A Noise-Embedded Federated Diffusion Model For Low-Count Whole-Body PET Denoising
Mar 20, 2025



Abstract:Low-count positron emission tomography (LCPET) imaging can reduce patients' exposure to radiation but often suffers from increased image noise and reduced lesion detectability, necessitating effective denoising techniques. Diffusion models have shown promise in LCPET denoising for recovering degraded image quality. However, training such models requires large and diverse datasets, which are challenging to obtain in the medical domain. To address data scarcity and privacy concerns, we combine diffusion models with federated learning -- a decentralized training approach where models are trained individually at different sites, and their parameters are aggregated on a central server over multiple iterations. The variation in scanner types and image noise levels within and across institutions poses additional challenges for federated learning in LCPET denoising. In this study, we propose a novel noise-embedded federated learning diffusion model (Fed-NDIF) to address these challenges, leveraging a multicenter dataset and varying count levels. Our approach incorporates liver normalized standard deviation (NSTD) noise embedding into a 2.5D diffusion model and utilizes the Federated Averaging (FedAvg) algorithm to aggregate locally trained models into a global model, which is subsequently fine-tuned on local datasets to optimize performance and obtain personalized models. Extensive validation on datasets from the University of Bern, Ruijin Hospital in Shanghai, and Yale-New Haven Hospital demonstrates the superior performance of our method in enhancing image quality and improving lesion quantification. The Fed-NDIF model shows significant improvements in PSNR, SSIM, and NMSE of the entire 3D volume, as well as enhanced lesion detectability and quantification, compared to local diffusion models and federated UNet-based models.
Anatomically and Metabolically Informed Diffusion for Unified Denoising and Segmentation in Low-Count PET Imaging
Mar 17, 2025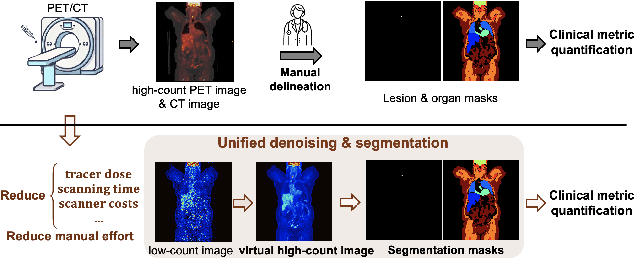
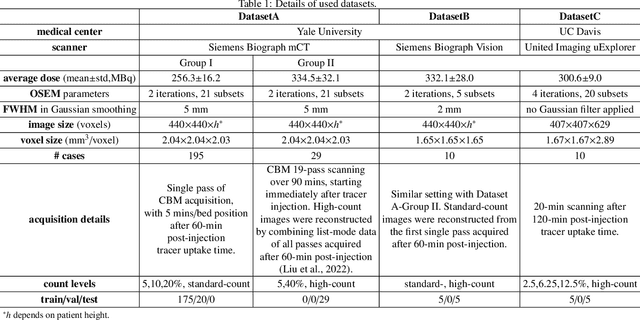
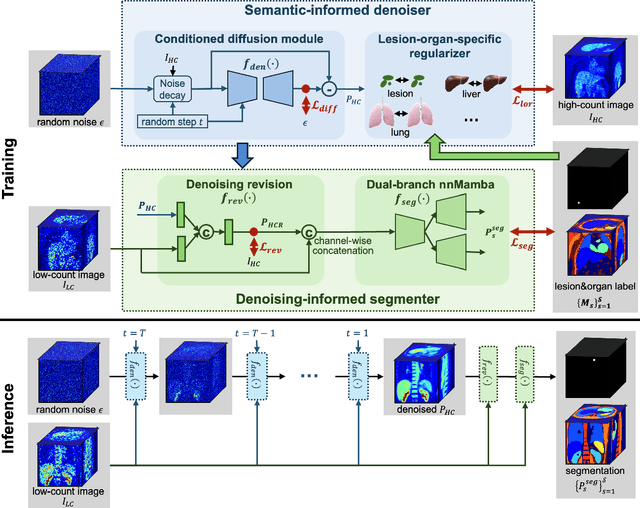
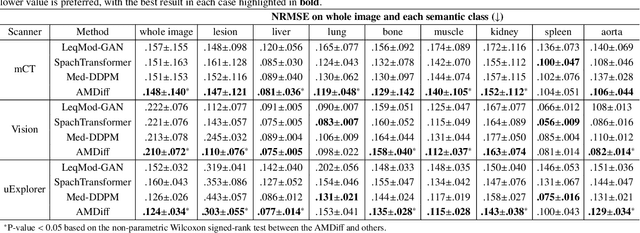
Abstract:Positron emission tomography (PET) image denoising, along with lesion and organ segmentation, are critical steps in PET-aided diagnosis. However, existing methods typically treat these tasks independently, overlooking inherent synergies between them as correlated steps in the analysis pipeline. In this work, we present the anatomically and metabolically informed diffusion (AMDiff) model, a unified framework for denoising and lesion/organ segmentation in low-count PET imaging. By integrating multi-task functionality and exploiting the mutual benefits of these tasks, AMDiff enables direct quantification of clinical metrics, such as total lesion glycolysis (TLG), from low-count inputs. The AMDiff model incorporates a semantic-informed denoiser based on diffusion strategy and a denoising-informed segmenter utilizing nnMamba architecture. The segmenter constrains denoised outputs via a lesion-organ-specific regularizer, while the denoiser enhances the segmenter by providing enriched image information through a denoising revision module. These components are connected via a warming-up mechanism to optimize multitask interactions. Experiments on multi-vendor, multi-center, and multi-noise-level datasets demonstrate the superior performance of AMDiff. For test cases below 20% of the clinical count levels from participating sites, AMDiff achieves TLG quantification biases of -26.98%, outperforming its ablated versions which yield biases of -35.85% (without the lesion-organ-specific regularizer) and -40.79% (without the denoising revision module).
A Generalizable 3D Diffusion Framework for Low-Dose and Few-View Cardiac SPECT
Dec 21, 2024Abstract:Myocardial perfusion imaging using SPECT is widely utilized to diagnose coronary artery diseases, but image quality can be negatively affected in low-dose and few-view acquisition settings. Although various deep learning methods have been introduced to improve image quality from low-dose or few-view SPECT data, previous approaches often fail to generalize across different acquisition settings, limiting their applicability in reality. This work introduced DiffSPECT-3D, a diffusion framework for 3D cardiac SPECT imaging that effectively adapts to different acquisition settings without requiring further network re-training or fine-tuning. Using both image and projection data, a consistency strategy is proposed to ensure that diffusion sampling at each step aligns with the low-dose/few-view projection measurements, the image data, and the scanner geometry, thus enabling generalization to different low-dose/few-view settings. Incorporating anatomical spatial information from CT and total variation constraint, we proposed a 2.5D conditional strategy to allow the DiffSPECT-3D to observe 3D contextual information from the entire image volume, addressing the 3D memory issues in diffusion model. We extensively evaluated the proposed method on 1,325 clinical 99mTc tetrofosmin stress/rest studies from 795 patients. Each study was reconstructed into 5 different low-count and 5 different few-view levels for model evaluations, ranging from 1% to 50% and from 1 view to 9 view, respectively. Validated against cardiac catheterization results and diagnostic comments from nuclear cardiologists, the presented results show the potential to achieve low-dose and few-view SPECT imaging without compromising clinical performance. Additionally, DiffSPECT-3D could be directly applied to full-dose SPECT images to further improve image quality, especially in a low-dose stress-first cardiac SPECT imaging protocol.
Noise-aware Dynamic Image Denoising and Positron Range Correction for Rubidium-82 Cardiac PET Imaging via Self-supervision
Sep 17, 2024



Abstract:Rb-82 is a radioactive isotope widely used for cardiac PET imaging. Despite numerous benefits of 82-Rb, there are several factors that limits its image quality and quantitative accuracy. First, the short half-life of 82-Rb results in noisy dynamic frames. Low signal-to-noise ratio would result in inaccurate and biased image quantification. Noisy dynamic frames also lead to highly noisy parametric images. The noise levels also vary substantially in different dynamic frames due to radiotracer decay and short half-life. Existing denoising methods are not applicable for this task due to the lack of paired training inputs/labels and inability to generalize across varying noise levels. Second, 82-Rb emits high-energy positrons. Compared with other tracers such as 18-F, 82-Rb travels a longer distance before annihilation, which negatively affect image spatial resolution. Here, the goal of this study is to propose a self-supervised method for simultaneous (1) noise-aware dynamic image denoising and (2) positron range correction for 82-Rb cardiac PET imaging. Tested on a series of PET scans from a cohort of normal volunteers, the proposed method produced images with superior visual quality. To demonstrate the improvement in image quantification, we compared image-derived input functions (IDIFs) with arterial input functions (AIFs) from continuous arterial blood samples. The IDIF derived from the proposed method led to lower AUC differences, decreasing from 11.09% to 7.58% on average, compared to the original dynamic frames. The proposed method also improved the quantification of myocardium blood flow (MBF), as validated against 15-O-water scans, with mean MBF differences decreased from 0.43 to 0.09, compared to the original dynamic frames. We also conducted a generalizability experiment on 37 patient scans obtained from a different country using a different scanner.
2.5D Multi-view Averaging Diffusion Model for 3D Medical Image Translation: Application to Low-count PET Reconstruction with CT-less Attenuation Correction
Jun 12, 2024
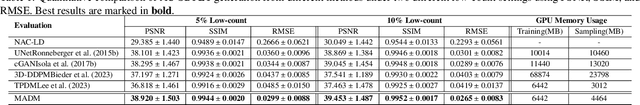

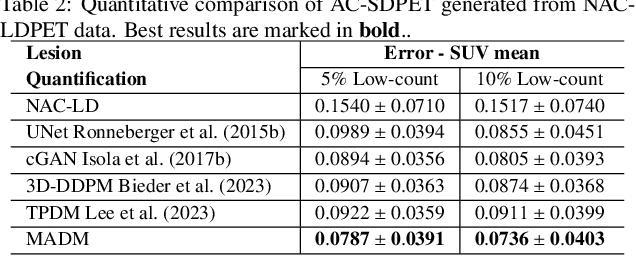
Abstract:Positron Emission Tomography (PET) is an important clinical imaging tool but inevitably introduces radiation hazards to patients and healthcare providers. Reducing the tracer injection dose and eliminating the CT acquisition for attenuation correction can reduce the overall radiation dose, but often results in PET with high noise and bias. Thus, it is desirable to develop 3D methods to translate the non-attenuation-corrected low-dose PET (NAC-LDPET) into attenuation-corrected standard-dose PET (AC-SDPET). Recently, diffusion models have emerged as a new state-of-the-art deep learning method for image-to-image translation, better than traditional CNN-based methods. However, due to the high computation cost and memory burden, it is largely limited to 2D applications. To address these challenges, we developed a novel 2.5D Multi-view Averaging Diffusion Model (MADM) for 3D image-to-image translation with application on NAC-LDPET to AC-SDPET translation. Specifically, MADM employs separate diffusion models for axial, coronal, and sagittal views, whose outputs are averaged in each sampling step to ensure the 3D generation quality from multiple views. To accelerate the 3D sampling process, we also proposed a strategy to use the CNN-based 3D generation as a prior for the diffusion model. Our experimental results on human patient studies suggested that MADM can generate high-quality 3D translation images, outperforming previous CNN-based and Diffusion-based baseline methods.
LpQcM: Adaptable Lesion-Quantification-Consistent Modulation for Deep Learning Low-Count PET Image Denoising
Apr 27, 2024Abstract:Deep learning-based positron emission tomography (PET) image denoising offers the potential to reduce radiation exposure and scanning time by transforming low-count images into high-count equivalents. However, existing methods typically blur crucial details, leading to inaccurate lesion quantification. This paper proposes a lesion-perceived and quantification-consistent modulation (LpQcM) strategy for enhanced PET image denoising, via employing downstream lesion quantification analysis as auxiliary tools. The LpQcM is a plug-and-play design adaptable to a wide range of model architectures, modulating the sampling and optimization procedures of model training without adding any computational burden to the inference phase. Specifically, the LpQcM consists of two components, the lesion-perceived modulation (LpM) and the multiscale quantification-consistent modulation (QcM). The LpM enhances lesion contrast and visibility by allocating higher sampling weights and stricter loss criteria to lesion-present samples determined by an auxiliary segmentation network than lesion-absent ones. The QcM further emphasizes accuracy of quantification for both the mean and maximum standardized uptake value (SUVmean and SUVmax) across multiscale sub-regions throughout the entire image, thereby enhancing the overall image quality. Experiments conducted on large PET datasets from multiple centers and vendors, and varying noise levels demonstrated the LpQcM efficacy across various denoising frameworks. Compared to frameworks without LpQcM, the integration of LpQcM reduces the lesion SUVmean bias by 2.92% on average and increases the peak signal-to-noise ratio (PSNR) by 0.34 on average, for denoising images of extremely low-count levels below 10%.
USFM: A Universal Ultrasound Foundation Model Generalized to Tasks and Organs towards Label Efficient Image Analysis
Jan 02, 2024Abstract:Inadequate generality across different organs and tasks constrains the application of ultrasound (US) image analysis methods in smart healthcare. Building a universal US foundation model holds the potential to address these issues. Nevertheless, the development of such foundational models encounters intrinsic challenges in US analysis, i.e., insufficient databases, low quality, and ineffective features. In this paper, we present a universal US foundation model, named USFM, generalized to diverse tasks and organs towards label efficient US image analysis. First, a large-scale Multi-organ, Multi-center, and Multi-device US database was built, comprehensively containing over two million US images. Organ-balanced sampling was employed for unbiased learning. Then, USFM is self-supervised pre-trained on the sufficient US database. To extract the effective features from low-quality US images, we proposed a spatial-frequency dual masked image modeling method. A productive spatial noise addition-recovery approach was designed to learn meaningful US information robustly, while a novel frequency band-stop masking learning approach was also employed to extract complex, implicit grayscale distribution and textural variations. Extensive experiments were conducted on the various tasks of segmentation, classification, and image enhancement from diverse organs and diseases. Comparisons with representative US image analysis models illustrate the universality and effectiveness of USFM. The label efficiency experiments suggest the USFM obtains robust performance with only 20% annotation, laying the groundwork for the rapid development of US models in clinical practices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge