Jinsong Ouyang
Dual Prompting for Diverse Count-level PET Denoising
May 05, 2025Abstract:The to-be-denoised positron emission tomography (PET) volumes are inherent with diverse count levels, which imposes challenges for a unified model to tackle varied cases. In this work, we resort to the recently flourished prompt learning to achieve generalizable PET denoising with different count levels. Specifically, we propose dual prompts to guide the PET denoising in a divide-and-conquer manner, i.e., an explicitly count-level prompt to provide the specific prior information and an implicitly general denoising prompt to encode the essential PET denoising knowledge. Then, a novel prompt fusion module is developed to unify the heterogeneous prompts, followed by a prompt-feature interaction module to inject prompts into the features. The prompts are able to dynamically guide the noise-conditioned denoising process. Therefore, we are able to efficiently train a unified denoising model for various count levels, and deploy it to different cases with personalized prompts. We evaluated on 1940 low-count PET 3D volumes with uniformly randomly selected 13-22\% fractions of events from 97 $^{18}$F-MK6240 tau PET studies. It shows our dual prompting can largely improve the performance with informed count-level and outperform the count-conditional model.
Mixture-of-Shape-Experts (MoSE): End-to-End Shape Dictionary Framework to Prompt SAM for Generalizable Medical Segmentation
Apr 13, 2025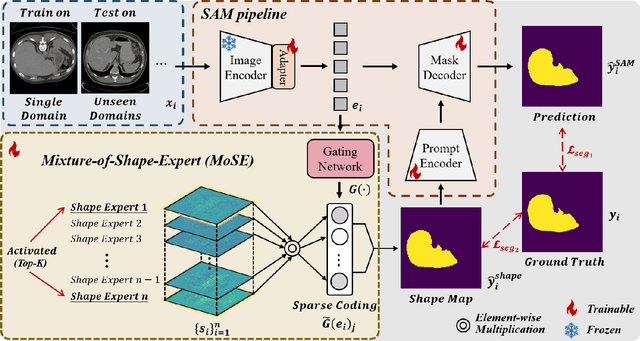
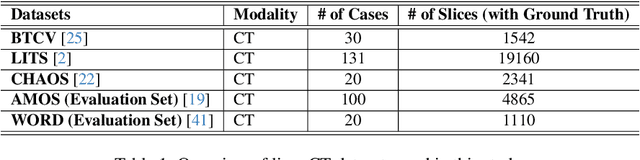
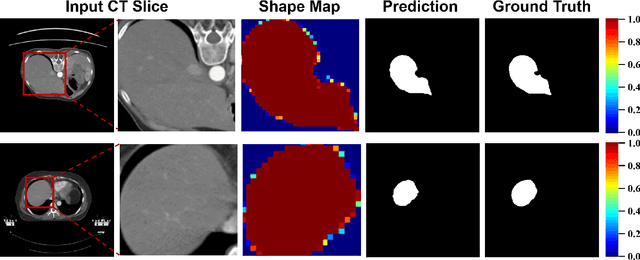
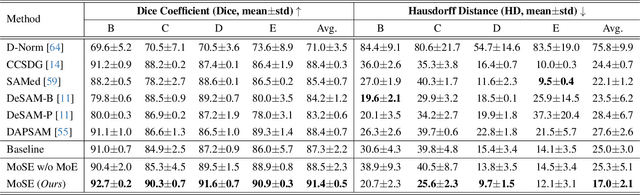
Abstract:Single domain generalization (SDG) has recently attracted growing attention in medical image segmentation. One promising strategy for SDG is to leverage consistent semantic shape priors across different imaging protocols, scanner vendors, and clinical sites. However, existing dictionary learning methods that encode shape priors often suffer from limited representational power with a small set of offline computed shape elements, or overfitting when the dictionary size grows. Moreover, they are not readily compatible with large foundation models such as the Segment Anything Model (SAM). In this paper, we propose a novel Mixture-of-Shape-Experts (MoSE) framework that seamlessly integrates the idea of mixture-of-experts (MoE) training into dictionary learning to efficiently capture diverse and robust shape priors. Our method conceptualizes each dictionary atom as a shape expert, which specializes in encoding distinct semantic shape information. A gating network dynamically fuses these shape experts into a robust shape map, with sparse activation guided by SAM encoding to prevent overfitting. We further provide this shape map as a prompt to SAM, utilizing the powerful generalization capability of SAM through bidirectional integration. All modules, including the shape dictionary, are trained in an end-to-end manner. Extensive experiments on multiple public datasets demonstrate its effectiveness.
Anatomically and Metabolically Informed Diffusion for Unified Denoising and Segmentation in Low-Count PET Imaging
Mar 17, 2025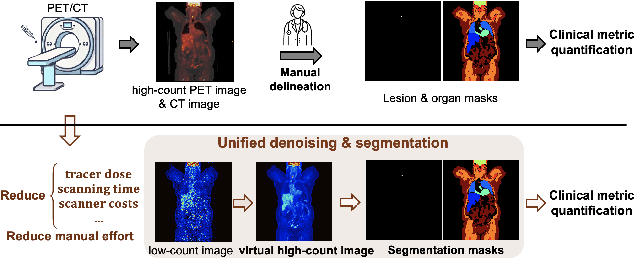
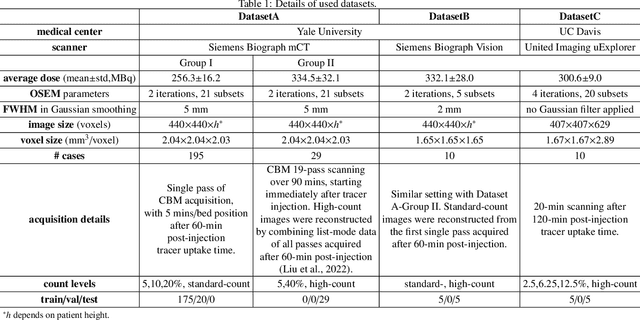
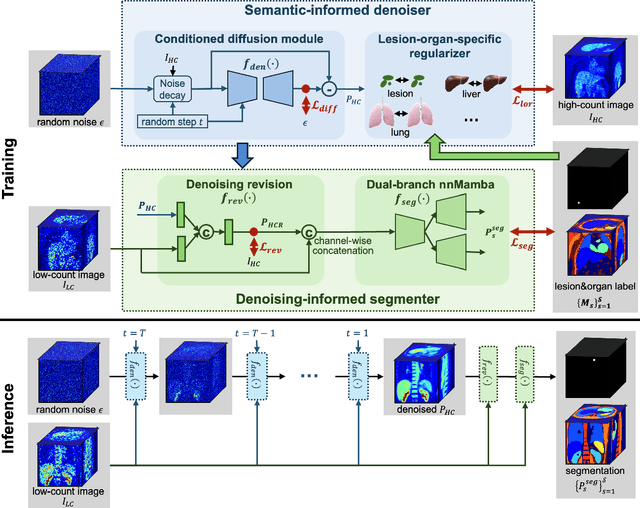
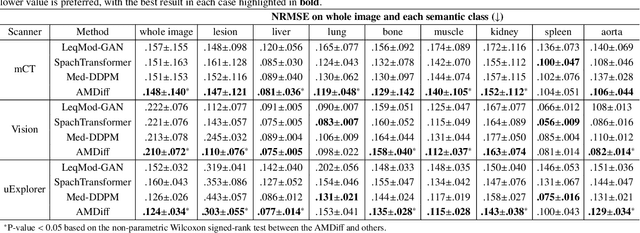
Abstract:Positron emission tomography (PET) image denoising, along with lesion and organ segmentation, are critical steps in PET-aided diagnosis. However, existing methods typically treat these tasks independently, overlooking inherent synergies between them as correlated steps in the analysis pipeline. In this work, we present the anatomically and metabolically informed diffusion (AMDiff) model, a unified framework for denoising and lesion/organ segmentation in low-count PET imaging. By integrating multi-task functionality and exploiting the mutual benefits of these tasks, AMDiff enables direct quantification of clinical metrics, such as total lesion glycolysis (TLG), from low-count inputs. The AMDiff model incorporates a semantic-informed denoiser based on diffusion strategy and a denoising-informed segmenter utilizing nnMamba architecture. The segmenter constrains denoised outputs via a lesion-organ-specific regularizer, while the denoiser enhances the segmenter by providing enriched image information through a denoising revision module. These components are connected via a warming-up mechanism to optimize multitask interactions. Experiments on multi-vendor, multi-center, and multi-noise-level datasets demonstrate the superior performance of AMDiff. For test cases below 20% of the clinical count levels from participating sites, AMDiff achieves TLG quantification biases of -26.98%, outperforming its ablated versions which yield biases of -35.85% (without the lesion-organ-specific regularizer) and -40.79% (without the denoising revision module).
Inspiring the Next Generation of Segment Anything Models: Comprehensively Evaluate SAM and SAM 2 with Diverse Prompts Towards Context-Dependent Concepts under Different Scenes
Dec 02, 2024Abstract:As a foundational model, SAM has significantly influenced multiple fields within computer vision, and its upgraded version, SAM 2, enhances capabilities in video segmentation, poised to make a substantial impact once again. While SAMs (SAM and SAM 2) have demonstrated excellent performance in segmenting context-independent concepts like people, cars, and roads, they overlook more challenging context-dependent (CD) concepts, such as visual saliency, camouflage, product defects, and medical lesions. CD concepts rely heavily on global and local contextual information, making them susceptible to shifts in different contexts, which requires strong discriminative capabilities from the model. The lack of comprehensive evaluation of SAMs limits understanding of their performance boundaries, which may hinder the design of future models. In this paper, we conduct a thorough quantitative evaluation of SAMs on 11 CD concepts across 2D and 3D images and videos in various visual modalities within natural, medical, and industrial scenes. We develop a unified evaluation framework for SAM and SAM 2 that supports manual, automatic, and intermediate self-prompting, aided by our specific prompt generation and interaction strategies. We further explore the potential of SAM 2 for in-context learning and introduce prompt robustness testing to simulate real-world imperfect prompts. Finally, we analyze the benefits and limitations of SAMs in understanding CD concepts and discuss their future development in segmentation tasks. This work aims to provide valuable insights to guide future research in both context-independent and context-dependent concepts segmentation, potentially informing the development of the next version - SAM 3.
Point-supervised Brain Tumor Segmentation with Box-prompted MedSAM
Aug 01, 2024

Abstract:Delineating lesions and anatomical structure is important for image-guided interventions. Point-supervised medical image segmentation (PSS) has great potential to alleviate costly expert delineation labeling. However, due to the lack of precise size and boundary guidance, the effectiveness of PSS often falls short of expectations. Although recent vision foundational models, such as the medical segment anything model (MedSAM), have made significant advancements in bounding-box-prompted segmentation, it is not straightforward to utilize point annotation, and is prone to semantic ambiguity. In this preliminary study, we introduce an iterative framework to facilitate semantic-aware point-supervised MedSAM. Specifically, the semantic box-prompt generator (SBPG) module has the capacity to convert the point input into potential pseudo bounding box suggestions, which are explicitly refined by the prototype-based semantic similarity. This is then succeeded by a prompt-guided spatial refinement (PGSR) module that harnesses the exceptional generalizability of MedSAM to infer the segmentation mask, which also updates the box proposal seed in SBPG. Performance can be progressively improved with adequate iterations. We conducted an evaluation on BraTS2018 for the segmentation of whole brain tumors and demonstrated its superior performance compared to traditional PSS methods and on par with box-supervised methods.
Disentangled Multimodal Brain MR Image Translation via Transformer-based Modality Infuser
Feb 01, 2024Abstract:Multimodal Magnetic Resonance (MR) Imaging plays a crucial role in disease diagnosis due to its ability to provide complementary information by analyzing a relationship between multimodal images on the same subject. Acquiring all MR modalities, however, can be expensive, and, during a scanning session, certain MR images may be missed depending on the study protocol. The typical solution would be to synthesize the missing modalities from the acquired images such as using generative adversarial networks (GANs). Yet, GANs constructed with convolutional neural networks (CNNs) are likely to suffer from a lack of global relationships and mechanisms to condition the desired modality. To address this, in this work, we propose a transformer-based modality infuser designed to synthesize multimodal brain MR images. In our method, we extract modality-agnostic features from the encoder and then transform them into modality-specific features using the modality infuser. Furthermore, the modality infuser captures long-range relationships among all brain structures, leading to the generation of more realistic images. We carried out experiments on the BraTS 2018 dataset, translating between four MR modalities, and our experimental results demonstrate the superiority of our proposed method in terms of synthesis quality. In addition, we conducted experiments on a brain tumor segmentation task and different conditioning methods.
Posterior Estimation for Dynamic PET imaging using Conditional Variational Inference
Oct 24, 2023Abstract:This work aims efficiently estimating the posterior distribution of kinetic parameters for dynamic positron emission tomography (PET) imaging given a measurement of time of activity curve. Considering the inherent information loss from parametric imaging to measurement space with the forward kinetic model, the inverse mapping is ambiguous. The conventional (but expensive) solution can be the Markov Chain Monte Carlo (MCMC) sampling, which is known to produce unbiased asymptotical estimation. We propose a deep-learning-based framework for efficient posterior estimation. Specifically, we counteract the information loss in the forward process by introducing latent variables. Then, we use a conditional variational autoencoder (CVAE) and optimize its evidence lower bound. The well-trained decoder is able to infer the posterior with a given measurement and the sampled latent variables following a simple multivariate Gaussian distribution. We validate our CVAE-based method using unbiased MCMC as the reference for low-dimensional data (a single brain region) with the simplified reference tissue model.
Posterior Estimation Using Deep Learning: A Simulation Study of Compartmental Modeling in Dynamic PET
Mar 17, 2023Abstract:Background: In medical imaging, images are usually treated as deterministic, while their uncertainties are largely underexplored. Purpose: This work aims at using deep learning to efficiently estimate posterior distributions of imaging parameters, which in turn can be used to derive the most probable parameters as well as their uncertainties. Methods: Our deep learning-based approaches are based on a variational Bayesian inference framework, which is implemented using two different deep neural networks based on conditional variational auto-encoder (CVAE), CVAE-dual-encoder and CVAE-dual-decoder. The conventional CVAE framework, i.e., CVAE-vanilla, can be regarded as a simplified case of these two neural networks. We applied these approaches to a simulation study of dynamic brain PET imaging using a reference region-based kinetic model. Results: In the simulation study, we estimated posterior distributions of PET kinetic parameters given a measurement of time-activity curve. Our proposed CVAE-dual-encoder and CVAE-dual-decoder yield results that are in good agreement with the asymptotically unbiased posterior distributions sampled by Markov Chain Monte Carlo (MCMC). The CVAE-vanilla can also be used for estimating posterior distributions, although it has an inferior performance to both CVAE-dual-encoder and CVAE-dual-decoder. Conclusions: We have evaluated the performance of our deep learning approaches for estimating posterior distributions in dynamic brain PET. Our deep learning approaches yield posterior distributions, which are in good agreement with unbiased distributions estimated by MCMC. All these neural networks have different characteristics and can be chosen by the user for specific applications. The proposed methods are general and can be adapted to other problems.
Domain Generalization under Conditional and Label Shifts via Variational Bayesian Inference
Jul 22, 2021



Abstract:In this work, we propose a domain generalization (DG) approach to learn on several labeled source domains and transfer knowledge to a target domain that is inaccessible in training. Considering the inherent conditional and label shifts, we would expect the alignment of $p(x|y)$ and $p(y)$. However, the widely used domain invariant feature learning (IFL) methods relies on aligning the marginal concept shift w.r.t. $p(x)$, which rests on an unrealistic assumption that $p(y)$ is invariant across domains. We thereby propose a novel variational Bayesian inference framework to enforce the conditional distribution alignment w.r.t. $p(x|y)$ via the prior distribution matching in a latent space, which also takes the marginal label shift w.r.t. $p(y)$ into consideration with the posterior alignment. Extensive experiments on various benchmarks demonstrate that our framework is robust to the label shift and the cross-domain accuracy is significantly improved, thereby achieving superior performance over the conventional IFL counterparts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge