Jihoon Cho
Volumetric Conditioning Module to Control Pretrained Diffusion Models for 3D Medical Images
Oct 29, 2024



Abstract:Spatial control methods using additional modules on pretrained diffusion models have gained attention for enabling conditional generation in natural images. These methods guide the generation process with new conditions while leveraging the capabilities of large models. They could be beneficial as training strategies in the context of 3D medical imaging, where training a diffusion model from scratch is challenging due to high computational costs and data scarcity. However, the potential application of spatial control methods with additional modules to 3D medical images has not yet been explored. In this paper, we present a tailored spatial control method for 3D medical images with a novel lightweight module, Volumetric Conditioning Module (VCM). Our VCM employs an asymmetric U-Net architecture to effectively encode complex information from various levels of 3D conditions, providing detailed guidance in image synthesis. To examine the applicability of spatial control methods and the effectiveness of VCM for 3D medical data, we conduct experiments under single- and multimodal conditions scenarios across a wide range of dataset sizes, from extremely small datasets with 10 samples to large datasets with 500 samples. The experimental results show that the VCM is effective for conditional generation and efficient in terms of requiring less training data and computational resources. We further investigate the potential applications for our spatial control method through axial super-resolution for medical images. Our code is available at \url{https://github.com/Ahn-Ssu/VCM}
Two-Stage Approach for Brain MR Image Synthesis: 2D Image Synthesis and 3D Refinement
Oct 14, 2024Abstract:Despite significant advancements in automatic brain tumor segmentation methods, their performance is not guaranteed when certain MR sequences are missing. Addressing this issue, it is crucial to synthesize the missing MR images that reflect the unique characteristics of the absent modality with precise tumor representation. Typically, MRI synthesis methods generate partial images rather than full-sized volumes due to computational constraints. This limitation can lead to a lack of comprehensive 3D volumetric information and result in image artifacts during the merging process. In this paper, we propose a two-stage approach that first synthesizes MR images from 2D slices using a novel intensity encoding method and then refines the synthesized MRI. The proposed intensity encoding reduces artifacts when synthesizing MRI on a 2D slice basis. Then, the \textit{Refiner}, which leverages complete 3D volume information, further improves the quality of the synthesized images and enhances their applicability to segmentation methods. Experimental results demonstrate that the intensity encoding effectively minimizes artifacts in the synthesized MRI and improves perceptual quality. Furthermore, using the \textit{Refiner} on synthesized MRI significantly improves brain tumor segmentation results, highlighting the potential of our approach in practical applications.
Label-Efficient 3D Brain Segmentation via Complementary 2D Diffusion Models with Orthogonal Views
Jul 17, 2024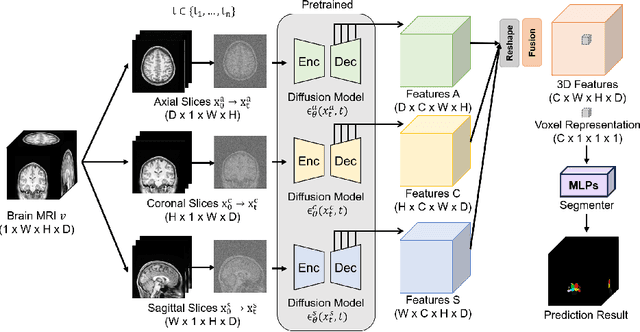
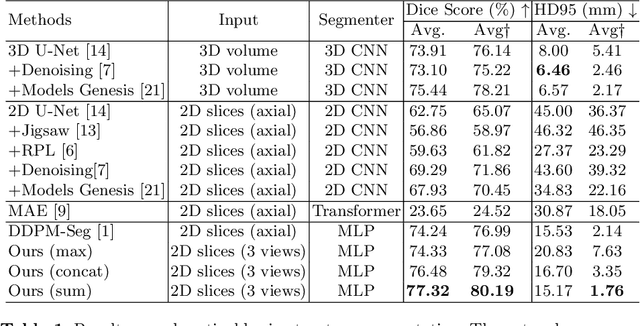
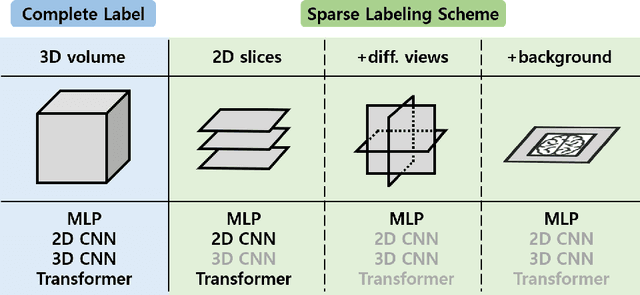
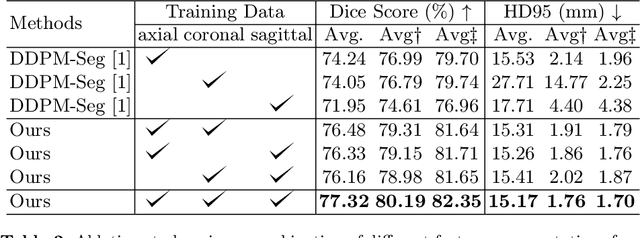
Abstract:Deep learning-based segmentation techniques have shown remarkable performance in brain segmentation, yet their success hinges on the availability of extensive labeled training data. Acquiring such vast datasets, however, poses a significant challenge in many clinical applications. To address this issue, in this work, we propose a novel 3D brain segmentation approach using complementary 2D diffusion models. The core idea behind our approach is to first mine 2D features with semantic information extracted from the 2D diffusion models by taking orthogonal views as input, followed by fusing them into a 3D contextual feature representation. Then, we use these aggregated features to train multi-layer perceptrons to classify the segmentation labels. Our goal is to achieve reliable segmentation quality without requiring complete labels for each individual subject. Our experiments on training in brain subcortical structure segmentation with a dataset from only one subject demonstrate that our approach outperforms state-of-the-art self-supervised learning methods. Further experiments on the minimum requirement of annotation by sparse labeling yield promising results even with only nine slices and a labeled background region.
A Unified Framework for Synthesizing Multisequence Brain MRI via Hybrid Fusion
Jun 21, 2024Abstract:Multisequence Magnetic Resonance Imaging (MRI) provides a reliable diagnosis in clinical applications through complementary information within sequences. However, in practice, the absence of certain MR sequences is a common problem that can lead to inconsistent analysis results. In this work, we propose a novel unified framework for synthesizing multisequence MR images, called Hybrid Fusion GAN (HF-GAN). We introduce a hybrid fusion encoder designed to ensure the disentangled extraction of complementary and modality-specific information, along with a channel attention-based feature fusion module that integrates the features into a common latent space handling the complexity from combinations of accessible MR sequences. Common feature representations are transformed into a target latent space via the modality infuser to synthesize missing MR sequences. We have performed experiments on multisequence brain MRI datasets from healthy individuals and patients diagnosed with brain tumors. Experimental results show that our method outperforms state-of-the-art methods in both quantitative and qualitative comparisons. In addition, a detailed analysis of our framework demonstrates the superiority of our designed modules and their effectiveness for use in data imputation tasks.
Disentangled Multimodal Brain MR Image Translation via Transformer-based Modality Infuser
Feb 01, 2024Abstract:Multimodal Magnetic Resonance (MR) Imaging plays a crucial role in disease diagnosis due to its ability to provide complementary information by analyzing a relationship between multimodal images on the same subject. Acquiring all MR modalities, however, can be expensive, and, during a scanning session, certain MR images may be missed depending on the study protocol. The typical solution would be to synthesize the missing modalities from the acquired images such as using generative adversarial networks (GANs). Yet, GANs constructed with convolutional neural networks (CNNs) are likely to suffer from a lack of global relationships and mechanisms to condition the desired modality. To address this, in this work, we propose a transformer-based modality infuser designed to synthesize multimodal brain MR images. In our method, we extract modality-agnostic features from the encoder and then transform them into modality-specific features using the modality infuser. Furthermore, the modality infuser captures long-range relationships among all brain structures, leading to the generation of more realistic images. We carried out experiments on the BraTS 2018 dataset, translating between four MR modalities, and our experimental results demonstrate the superiority of our proposed method in terms of synthesis quality. In addition, we conducted experiments on a brain tumor segmentation task and different conditioning methods.
Hybrid-Fusion Transformer for Multisequence MRI
Nov 02, 2023Abstract:Medical segmentation has grown exponentially through the advent of a fully convolutional network (FCN), and we have now reached a turning point through the success of Transformer. However, the different characteristics of the modality have not been fully integrated into Transformer for medical segmentation. In this work, we propose the novel hybrid fusion Transformer (HFTrans) for multisequence MRI image segmentation. We take advantage of the differences among multimodal MRI sequences and utilize the Transformer layers to integrate the features extracted from each modality as well as the features of the early fused modalities. We validate the effectiveness of our hybrid-fusion method in three-dimensional (3D) medical segmentation. Experiments on two public datasets, BraTS2020 and MRBrainS18, show that the proposed method outperforms previous state-of-the-art methods on the task of brain tumor segmentation and brain structure segmentation.
Why is the winner the best?
Mar 30, 2023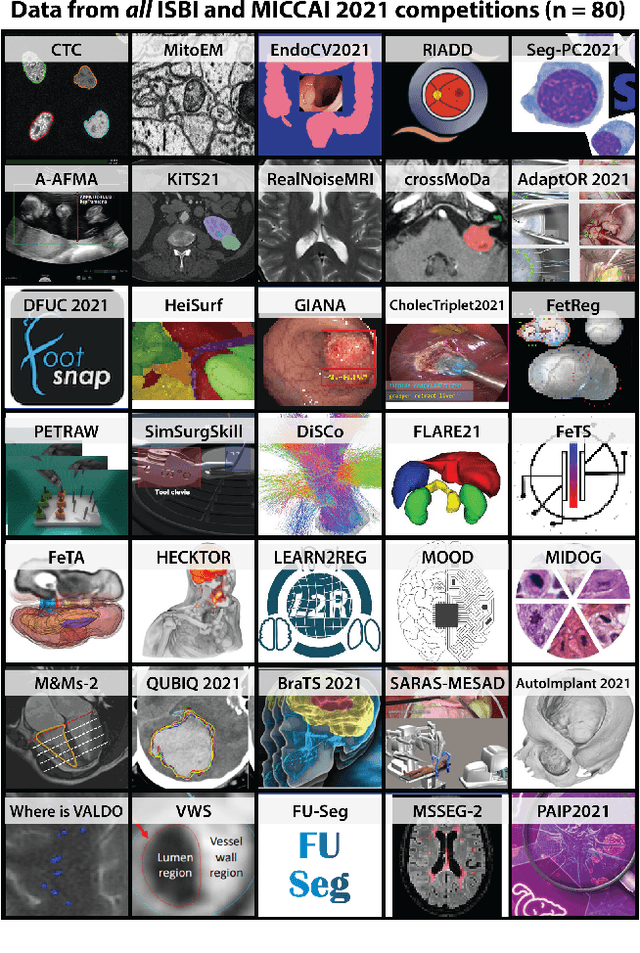
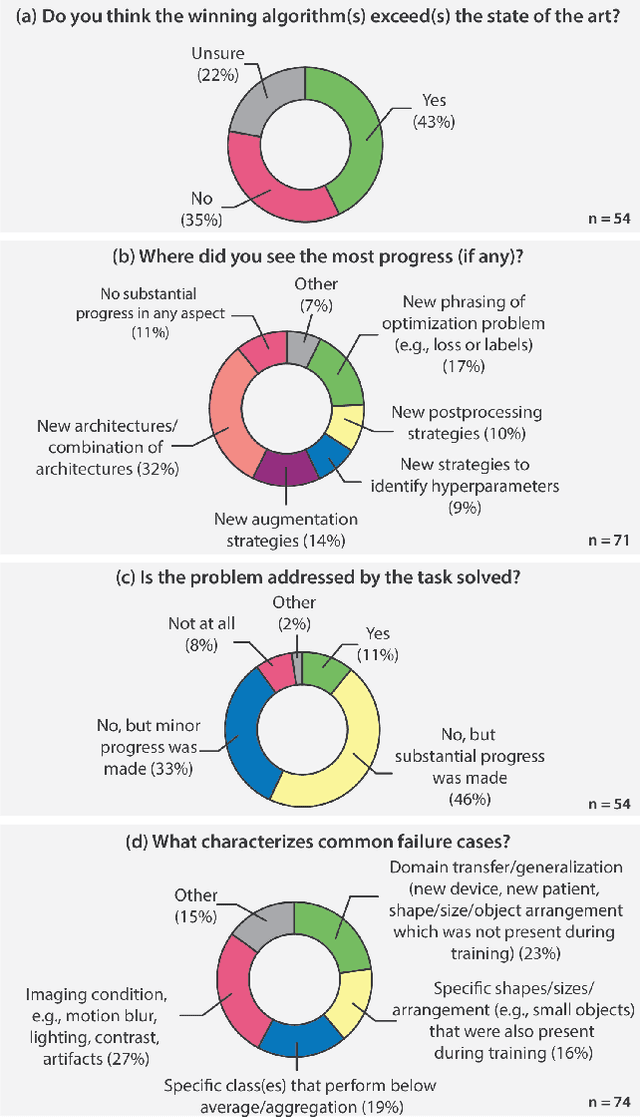
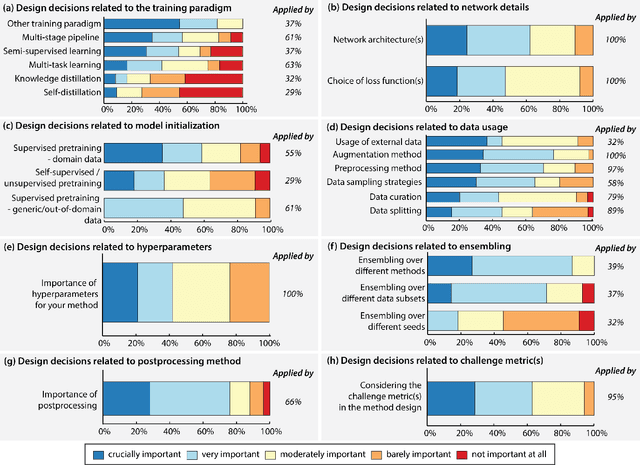
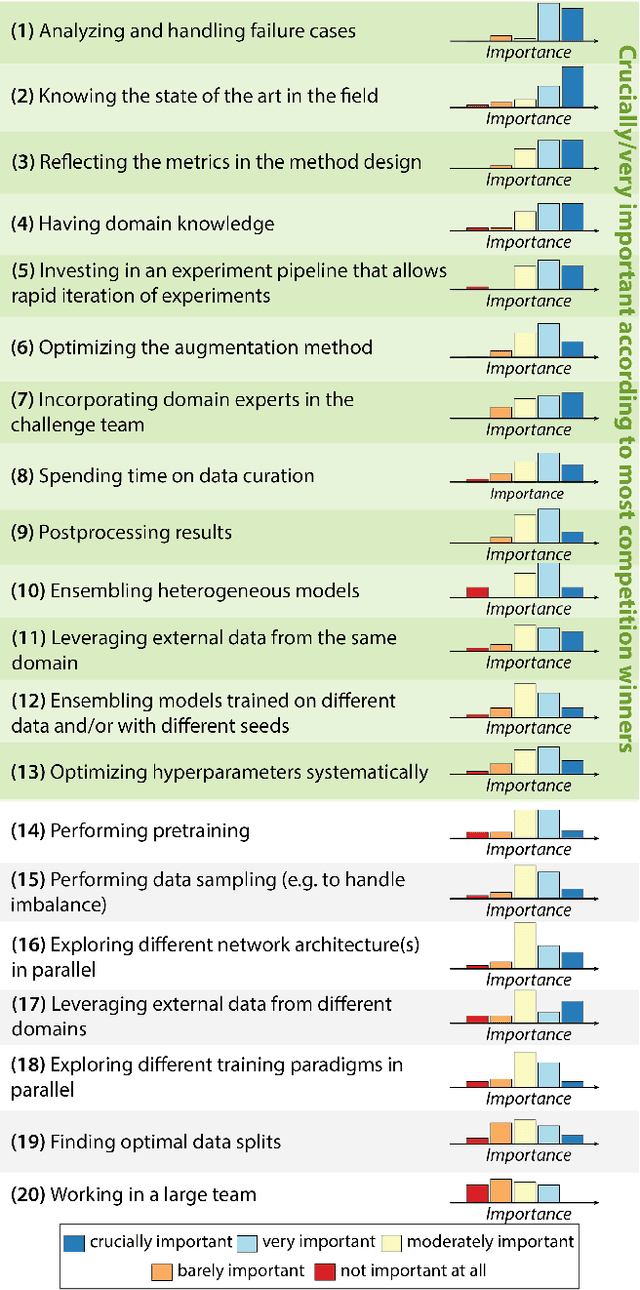
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
SOS: Score-based Oversampling for Tabular Data
Jun 17, 2022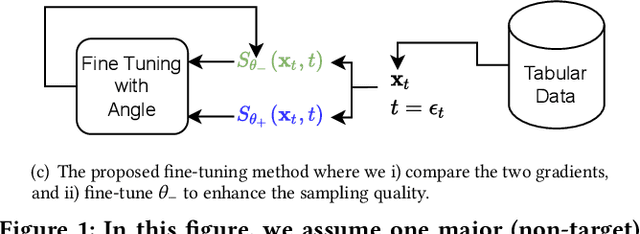
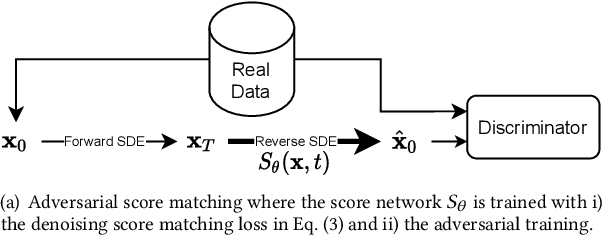
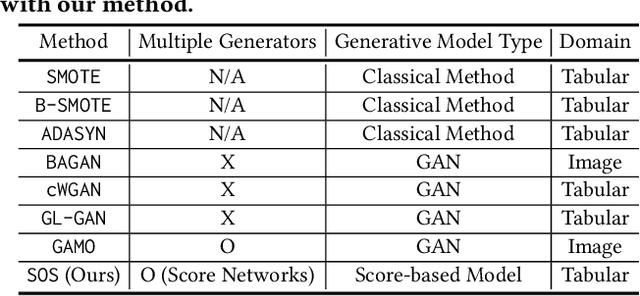
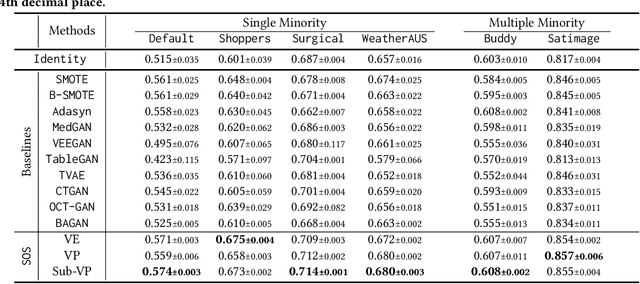
Abstract:Score-based generative models (SGMs) are a recent breakthrough in generating fake images. SGMs are known to surpass other generative models, e.g., generative adversarial networks (GANs) and variational autoencoders (VAEs). Being inspired by their big success, in this work, we fully customize them for generating fake tabular data. In particular, we are interested in oversampling minor classes since imbalanced classes frequently lead to sub-optimal training outcomes. To our knowledge, we are the first presenting a score-based tabular data oversampling method. Firstly, we re-design our own score network since we have to process tabular data. Secondly, we propose two options for our generation method: the former is equivalent to a style transfer for tabular data and the latter uses the standard generative policy of SGMs. Lastly, we define a fine-tuning method, which further enhances the oversampling quality. In our experiments with 6 datasets and 10 baselines, our method outperforms other oversampling methods in all cases.
Invertible Tabular GANs: Killing Two Birds with OneStone for Tabular Data Synthesis
Feb 08, 2022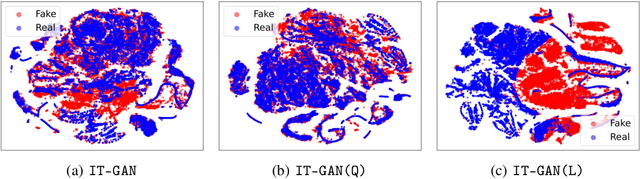
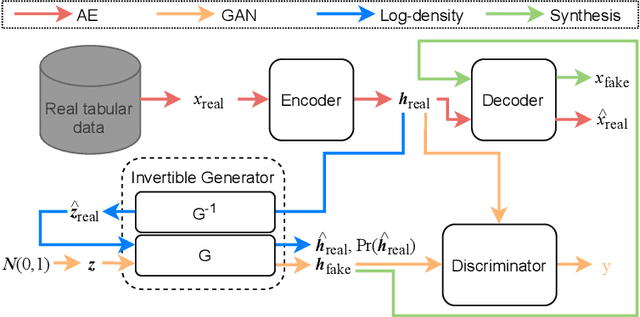
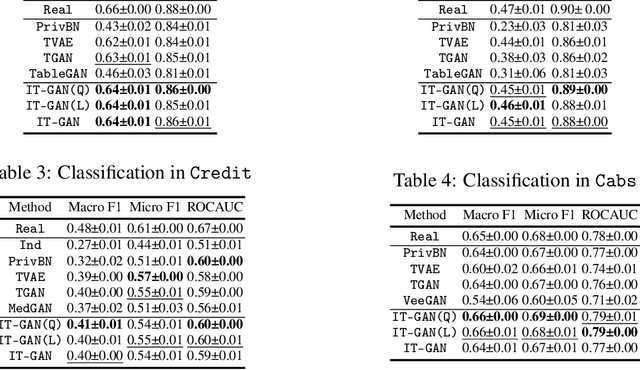
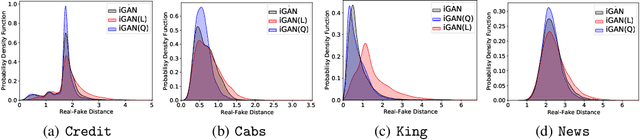
Abstract:Tabular data synthesis has received wide attention in the literature. This is because available data is often limited, incomplete, or cannot be obtained easily, and data privacy is becoming increasingly important. In this work, we present a generalized GAN framework for tabular synthesis, which combines the adversarial training of GANs and the negative log-density regularization of invertible neural networks. The proposed framework can be used for two distinctive objectives. First, we can further improve the synthesis quality, by decreasing the negative log-density of real records in the process of adversarial training. On the other hand, by increasing the negative log-density of real records, realistic fake records can be synthesized in a way that they are not too much close to real records and reduce the chance of potential information leakage. We conduct experiments with real-world datasets for classification, regression, and privacy attacks. In general, the proposed method demonstrates the best synthesis quality (in terms of task-oriented evaluation metrics, e.g., F1) when decreasing the negative log-density during the adversarial training. If increasing the negative log-density, our experimental results show that the distance between real and fake records increases, enhancing robustness against privacy attacks.
Domain-Robust Mitotic Figure Detection with Style Transfer
Sep 30, 2021


Abstract:We propose a new training scheme for domain generalization in mitotic figure detection. Mitotic figures show different characteristics for each scanner. We consider each scanner as a 'domain' and the image distribution specified for each domain as 'style'. The goal is to train our network to be robust on scanner types by using various 'style' images. To expand the style variance, we transfer a style of the training image into arbitrary styles, by defining a module based on StarGAN. Our model with the proposed training scheme shows positive performance on MIDOG Preliminary Test-Set containing scanners never seen before.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge