Jonghye Woo
Interpretable and backpropagation-free Green Learning for efficient multi-task echocardiographic segmentation and classification
Jan 27, 2026Abstract:Echocardiography is a cornerstone for managing heart failure (HF), with Left Ventricular Ejection Fraction (LVEF) being a critical metric for guiding therapy. However, manual LVEF assessment suffers from high inter-observer variability, while existing Deep Learning (DL) models are often computationally intensive and data-hungry "black boxes" that impede clinical trust and adoption. Here, we propose a backpropagation-free multi-task Green Learning (MTGL) framework that performs simultaneous Left Ventricle (LV) segmentation and LVEF classification. Our framework integrates an unsupervised VoxelHop encoder for hierarchical spatio-temporal feature extraction with a multi-level regression decoder and an XG-Boost classifier. On the EchoNet-Dynamic dataset, our MTGL model achieves state-of-the-art classification and segmentation performance, attaining a classification accuracy of 94.3% and a Dice Similarity Coefficient (DSC) of 0.912, significantly outperforming several advanced 3D DL models. Crucially, our model achieves this with over an order of magnitude fewer parameters, demonstrating exceptional computational efficiency. This work demonstrates that the GL paradigm can deliver highly accurate, efficient, and interpretable solutions for complex medical image analysis, paving the way for more sustainable and trustworthy artificial intelligence in clinical practice.
Cross-Modal Fine-Tuning of 3D Convolutional Foundation Models for ADHD Classification with Low-Rank Adaptation
Nov 08, 2025Abstract:Early diagnosis of attention-deficit/hyperactivity disorder (ADHD) in children plays a crucial role in improving outcomes in education and mental health. Diagnosing ADHD using neuroimaging data, however, remains challenging due to heterogeneous presentations and overlapping symptoms with other conditions. To address this, we propose a novel parameter-efficient transfer learning approach that adapts a large-scale 3D convolutional foundation model, pre-trained on CT images, to an MRI-based ADHD classification task. Our method introduces Low-Rank Adaptation (LoRA) in 3D by factorizing 3D convolutional kernels into 2D low-rank updates, dramatically reducing trainable parameters while achieving superior performance. In a five-fold cross-validated evaluation on a public diffusion MRI database, our 3D LoRA fine-tuning strategy achieved state-of-the-art results, with one model variant reaching 71.9% accuracy and another attaining an AUC of 0.716. Both variants use only 1.64 million trainable parameters (over 113x fewer than a fully fine-tuned foundation model). Our results represent one of the first successful cross-modal (CT-to-MRI) adaptations of a foundation model in neuroimaging, establishing a new benchmark for ADHD classification while greatly improving efficiency.
Brightness-Invariant Tracking Estimation in Tagged MRI
May 23, 2025Abstract:Magnetic resonance (MR) tagging is an imaging technique for noninvasively tracking tissue motion in vivo by creating a visible pattern of magnetization saturation (tags) that deforms with the tissue. Due to longitudinal relaxation and progression to steady-state, the tags and tissue brightnesses change over time, which makes tracking with optical flow methods error-prone. Although Fourier methods can alleviate these problems, they are also sensitive to brightness changes as well as spectral spreading due to motion. To address these problems, we introduce the brightness-invariant tracking estimation (BRITE) technique for tagged MRI. BRITE disentangles the anatomy from the tag pattern in the observed tagged image sequence and simultaneously estimates the Lagrangian motion. The inherent ill-posedness of this problem is addressed by leveraging the expressive power of denoising diffusion probabilistic models to represent the probabilistic distribution of the underlying anatomy and the flexibility of physics-informed neural networks to estimate biologically-plausible motion. A set of tagged MR images of a gel phantom was acquired with various tag periods and imaging flip angles to demonstrate the impact of brightness variations and to validate our method. The results show that BRITE achieves more accurate motion and strain estimates as compared to other state of the art methods, while also being resistant to tag fading.
Mixture-of-Shape-Experts (MoSE): End-to-End Shape Dictionary Framework to Prompt SAM for Generalizable Medical Segmentation
Apr 13, 2025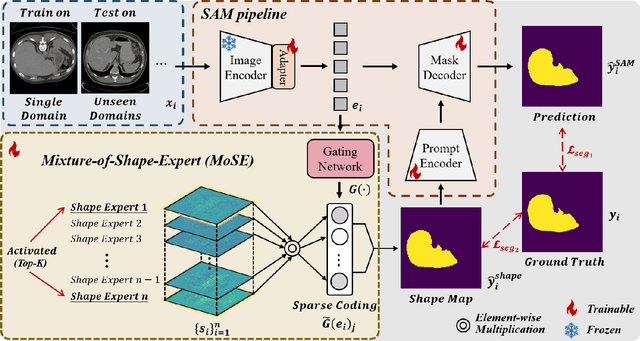
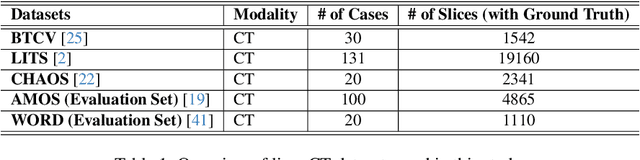
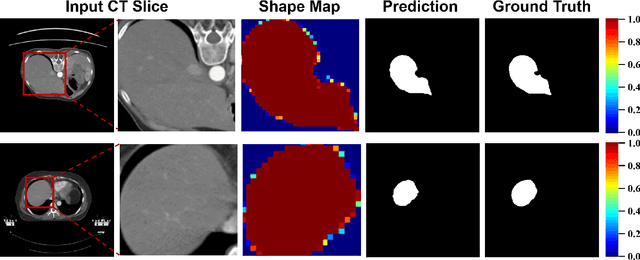
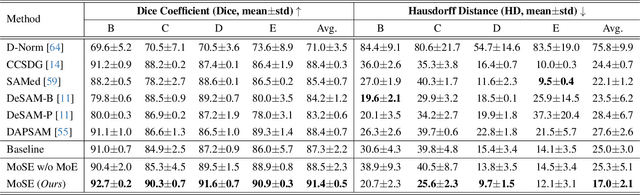
Abstract:Single domain generalization (SDG) has recently attracted growing attention in medical image segmentation. One promising strategy for SDG is to leverage consistent semantic shape priors across different imaging protocols, scanner vendors, and clinical sites. However, existing dictionary learning methods that encode shape priors often suffer from limited representational power with a small set of offline computed shape elements, or overfitting when the dictionary size grows. Moreover, they are not readily compatible with large foundation models such as the Segment Anything Model (SAM). In this paper, we propose a novel Mixture-of-Shape-Experts (MoSE) framework that seamlessly integrates the idea of mixture-of-experts (MoE) training into dictionary learning to efficiently capture diverse and robust shape priors. Our method conceptualizes each dictionary atom as a shape expert, which specializes in encoding distinct semantic shape information. A gating network dynamically fuses these shape experts into a robust shape map, with sparse activation guided by SAM encoding to prevent overfitting. We further provide this shape map as a prompt to SAM, utilizing the powerful generalization capability of SAM through bidirectional integration. All modules, including the shape dictionary, are trained in an end-to-end manner. Extensive experiments on multiple public datasets demonstrate its effectiveness.
A Speech-to-Video Synthesis Approach Using Spatio-Temporal Diffusion for Vocal Tract MRI
Mar 15, 2025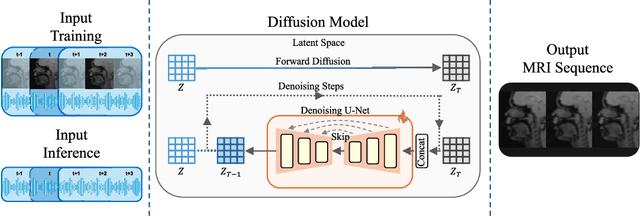
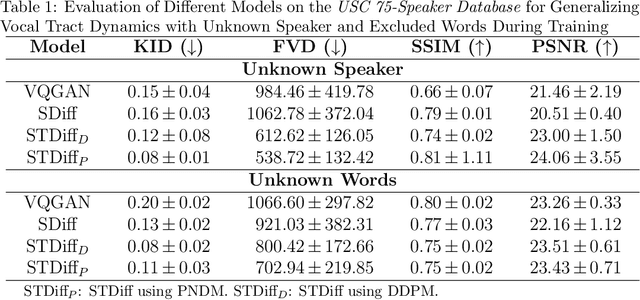
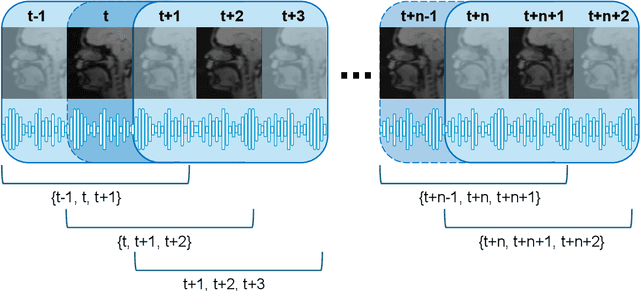

Abstract:Understanding the relationship between vocal tract motion during speech and the resulting acoustic signal is crucial for aided clinical assessment and developing personalized treatment and rehabilitation strategies. Toward this goal, we introduce an audio-to-video generation framework for creating Real Time/cine-Magnetic Resonance Imaging (RT-/cine-MRI) visuals of the vocal tract from speech signals. Our framework first preprocesses RT-/cine-MRI sequences and speech samples to achieve temporal alignment, ensuring synchronization between visual and audio data. We then employ a modified stable diffusion model, integrating structural and temporal blocks, to effectively capture movement characteristics and temporal dynamics in the synchronized data. This process enables the generation of MRI sequences from new speech inputs, improving the conversion of audio into visual data. We evaluated our framework on healthy controls and tongue cancer patients by analyzing and comparing the vocal tract movements in synthesized videos. Our framework demonstrated adaptability to new speech inputs and effective generalization. In addition, positive human evaluations confirmed its effectiveness, with realistic and accurate visualizations, suggesting its potential for outpatient therapy and personalized simulation of vocal tract visualizations.
Semi-Supervised Bone Marrow Lesion Detection from Knee MRI Segmentation Using Mask Inpainting Models
Sep 27, 2024



Abstract:Bone marrow lesions (BMLs) are critical indicators of knee osteoarthritis (OA). Since they often appear as small, irregular structures with indistinguishable edges in knee magnetic resonance images (MRIs), effective detection of BMLs in MRI is vital for OA diagnosis and treatment. This paper proposes a semi-supervised local anomaly detection method using mask inpainting models for identification of BMLs in high-resolution knee MRI, effectively integrating a 3D femur bone segmentation model, a large mask inpainting model, and a series of post-processing techniques. The method was evaluated using MRIs at various resolutions from a subset of the public Osteoarthritis Initiative database. Dice score, Intersection over Union (IoU), and pixel-level sensitivity, specificity, and accuracy showed an advantage over the multiresolution knowledge distillation method-a state-of-the-art global anomaly detection method. Especially, segmentation performance is enhanced on higher-resolution images, achieving an over two times performance increase on the Dice score and the IoU score at a 448x448 resolution level. We also demonstrate that with increasing size of the BML region, both the Dice and IoU scores improve as the proportion of distinguishable boundary decreases. The identified BML masks can serve as markers for downstream tasks such as segmentation and classification. The proposed method has shown a potential in improving BML detection, laying a foundation for further advances in imaging-based OA research.
Point-supervised Brain Tumor Segmentation with Box-prompted MedSAM
Aug 01, 2024

Abstract:Delineating lesions and anatomical structure is important for image-guided interventions. Point-supervised medical image segmentation (PSS) has great potential to alleviate costly expert delineation labeling. However, due to the lack of precise size and boundary guidance, the effectiveness of PSS often falls short of expectations. Although recent vision foundational models, such as the medical segment anything model (MedSAM), have made significant advancements in bounding-box-prompted segmentation, it is not straightforward to utilize point annotation, and is prone to semantic ambiguity. In this preliminary study, we introduce an iterative framework to facilitate semantic-aware point-supervised MedSAM. Specifically, the semantic box-prompt generator (SBPG) module has the capacity to convert the point input into potential pseudo bounding box suggestions, which are explicitly refined by the prototype-based semantic similarity. This is then succeeded by a prompt-guided spatial refinement (PGSR) module that harnesses the exceptional generalizability of MedSAM to infer the segmentation mask, which also updates the box proposal seed in SBPG. Performance can be progressively improved with adequate iterations. We conducted an evaluation on BraTS2018 for the segmentation of whole brain tumors and demonstrated its superior performance compared to traditional PSS methods and on par with box-supervised methods.
Label-Efficient 3D Brain Segmentation via Complementary 2D Diffusion Models with Orthogonal Views
Jul 17, 2024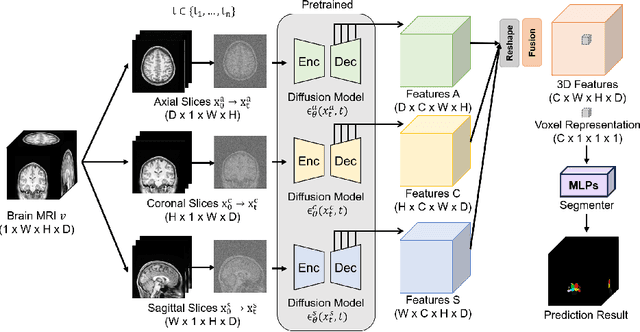
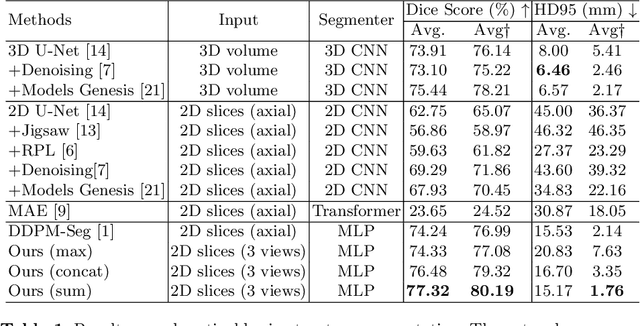
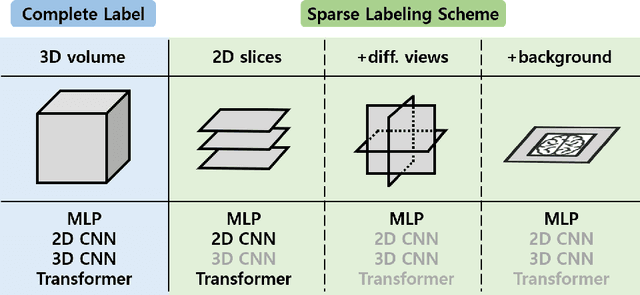
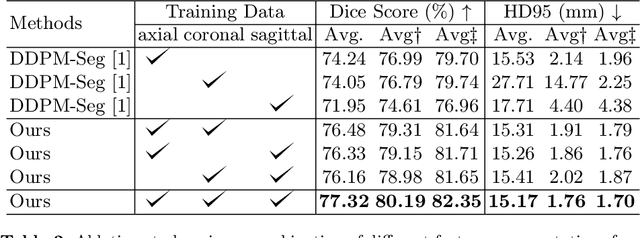
Abstract:Deep learning-based segmentation techniques have shown remarkable performance in brain segmentation, yet their success hinges on the availability of extensive labeled training data. Acquiring such vast datasets, however, poses a significant challenge in many clinical applications. To address this issue, in this work, we propose a novel 3D brain segmentation approach using complementary 2D diffusion models. The core idea behind our approach is to first mine 2D features with semantic information extracted from the 2D diffusion models by taking orthogonal views as input, followed by fusing them into a 3D contextual feature representation. Then, we use these aggregated features to train multi-layer perceptrons to classify the segmentation labels. Our goal is to achieve reliable segmentation quality without requiring complete labels for each individual subject. Our experiments on training in brain subcortical structure segmentation with a dataset from only one subject demonstrate that our approach outperforms state-of-the-art self-supervised learning methods. Further experiments on the minimum requirement of annotation by sparse labeling yield promising results even with only nine slices and a labeled background region.
A Unified Framework for Synthesizing Multisequence Brain MRI via Hybrid Fusion
Jun 21, 2024Abstract:Multisequence Magnetic Resonance Imaging (MRI) provides a reliable diagnosis in clinical applications through complementary information within sequences. However, in practice, the absence of certain MR sequences is a common problem that can lead to inconsistent analysis results. In this work, we propose a novel unified framework for synthesizing multisequence MR images, called Hybrid Fusion GAN (HF-GAN). We introduce a hybrid fusion encoder designed to ensure the disentangled extraction of complementary and modality-specific information, along with a channel attention-based feature fusion module that integrates the features into a common latent space handling the complexity from combinations of accessible MR sequences. Common feature representations are transformed into a target latent space via the modality infuser to synthesize missing MR sequences. We have performed experiments on multisequence brain MRI datasets from healthy individuals and patients diagnosed with brain tumors. Experimental results show that our method outperforms state-of-the-art methods in both quantitative and qualitative comparisons. In addition, a detailed analysis of our framework demonstrates the superiority of our designed modules and their effectiveness for use in data imputation tasks.
Treatment-wise Glioblastoma Survival Inference with Multi-parametric Preoperative MRI
Feb 10, 2024Abstract:In this work, we aim to predict the survival time (ST) of glioblastoma (GBM) patients undergoing different treatments based on preoperative magnetic resonance (MR) scans. The personalized and precise treatment planning can be achieved by comparing the ST of different treatments. It is well established that both the current status of the patient (as represented by the MR scans) and the choice of treatment are the cause of ST. While previous related MR-based glioblastoma ST studies have focused only on the direct mapping of MR scans to ST, they have not included the underlying causal relationship between treatments and ST. To address this limitation, we propose a treatment-conditioned regression model for glioblastoma ST that incorporates treatment information in addition to MR scans. Our approach allows us to effectively utilize the data from all of the treatments in a unified manner, rather than having to train separate models for each of the treatments. Furthermore, treatment can be effectively injected into each convolutional layer through the adaptive instance normalization we employ. We evaluate our framework on the BraTS20 ST prediction task. Three treatment options are considered: Gross Total Resection (GTR), Subtotal Resection (STR), and no resection. The evaluation results demonstrate the effectiveness of injecting the treatment for estimating GBM survival.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge