Samuel W. Remedios
Likelihood-Separable Diffusion Inference for Multi-Image MRI Super-Resolution
Jan 20, 2026Abstract:Diffusion models are the current state-of-the-art for solving inverse problems in imaging. Their impressive generative capability allows them to approximate sampling from a prior distribution, which alongside a known likelihood function permits posterior sampling without retraining the model. While recent methods have made strides in advancing the accuracy of posterior sampling, the majority focuses on single-image inverse problems. However, for modalities such as magnetic resonance imaging (MRI), it is common to acquire multiple complementary measurements, each low-resolution along a different axis. In this work, we generalize common diffusion-based inverse single-image problem solvers for multi-image super-resolution (MISR) MRI. We show that the DPS likelihood correction allows an exactly-separable gradient decomposition across independently acquired measurements, enabling MISR without constructing a joint operator, modifying the diffusion model, or increasing network function evaluations. We derive MISR versions of DPS, DMAP, DPPS, and diffusion-based PnP/ADMM, and demonstrate substantial gains over SISR across $4\times/8\times/16\times$ anisotropic degradations. Our results achieve state-of-the-art super-resolution of anisotropic MRI volumes and, critically, enable reconstruction of near-isotropic anatomy from routine 2D multi-slice acquisitions, which are otherwise highly degraded in orthogonal views.
MSRepaint: Multiple Sclerosis Repaint with Conditional Denoising Diffusion Implicit Model for Bidirectional Lesion Filling and Synthesis
Oct 02, 2025Abstract:In multiple sclerosis, lesions interfere with automated magnetic resonance imaging analyses such as brain parcellation and deformable registration, while lesion segmentation models are hindered by the limited availability of annotated training data. To address both issues, we propose MSRepaint, a unified diffusion-based generative model for bidirectional lesion filling and synthesis that restores anatomical continuity for downstream analyses and augments segmentation through realistic data generation. MSRepaint conditions on spatial lesion masks for voxel-level control, incorporates contrast dropout to handle missing inputs, integrates a repainting mechanism to preserve surrounding anatomy during lesion filling and synthesis, and employs a multi-view DDIM inversion and fusion pipeline for 3D consistency with fast inference. Extensive evaluations demonstrate the effectiveness of MSRepaint across multiple tasks. For lesion filling, we evaluate both the accuracy within the filled regions and the impact on downstream tasks including brain parcellation and deformable registration. MSRepaint outperforms the traditional lesion filling methods FSL and NiftySeg, and achieves accuracy on par with FastSurfer-LIT, a recent diffusion model-based inpainting method, while offering over 20 times faster inference. For lesion synthesis, state-of-the-art MS lesion segmentation models trained on MSRepaint-synthesized data outperform those trained on CarveMix-synthesized data or real ISBI challenge training data across multiple benchmarks, including the MICCAI 2016 and UMCL datasets. Additionally, we demonstrate that MSRepaint's unified bidirectional filling and synthesis capability, with full spatial control over lesion appearance, enables high-fidelity simulation of lesion evolution in longitudinal MS progression.
Segmenting Thalamic Nuclei: T1 Maps Provide a Reliable and Efficient Solution
Aug 17, 2025Abstract:Accurate thalamic nuclei segmentation is crucial for understanding neurological diseases, brain functions, and guiding clinical interventions. However, the optimal inputs for segmentation remain unclear. This study systematically evaluates multiple MRI contrasts, including MPRAGE and FGATIR sequences, quantitative PD and T1 maps, and multiple T1-weighted images at different inversion times (multi-TI), to determine the most effective inputs. For multi-TI images, we employ a gradient-based saliency analysis with Monte Carlo dropout and propose an Overall Importance Score to select the images contributing most to segmentation. A 3D U-Net is trained on each of these configurations. Results show that T1 maps alone achieve strong quantitative performance and superior qualitative outcomes, while PD maps offer no added value. These findings underscore the value of T1 maps as a reliable and efficient input among the evaluated options, providing valuable guidance for optimizing imaging protocols when thalamic structures are of clinical or research interest.
UNISELF: A Unified Network with Instance Normalization and Self-Ensembled Lesion Fusion for Multiple Sclerosis Lesion Segmentation
Aug 06, 2025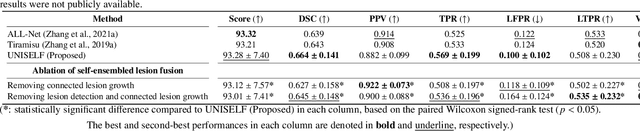
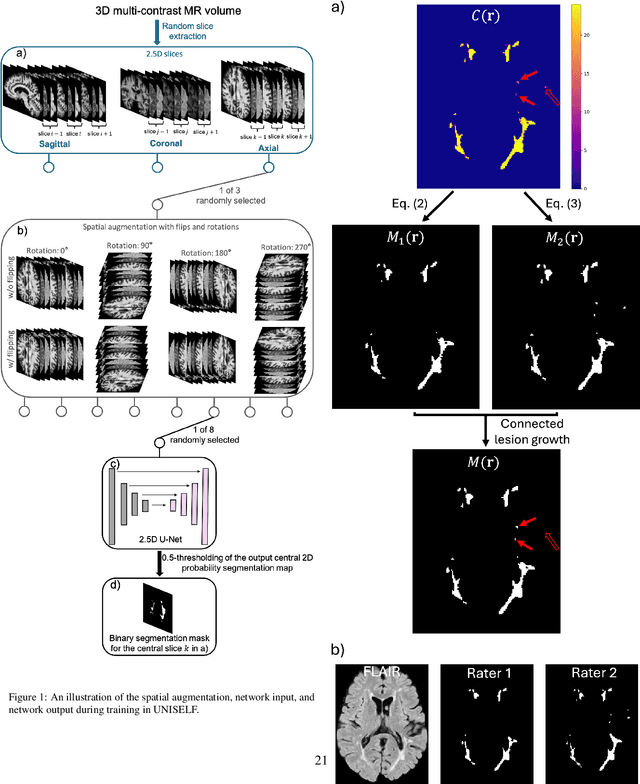
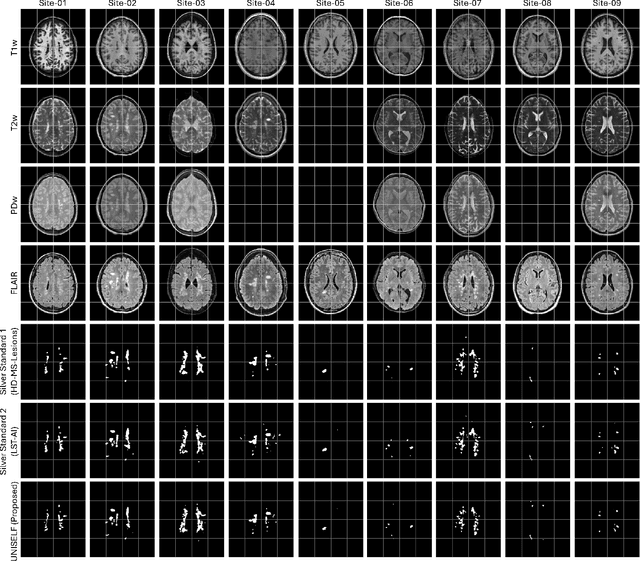

Abstract:Automated segmentation of multiple sclerosis (MS) lesions using multicontrast magnetic resonance (MR) images improves efficiency and reproducibility compared to manual delineation, with deep learning (DL) methods achieving state-of-the-art performance. However, these DL-based methods have yet to simultaneously optimize in-domain accuracy and out-of-domain generalization when trained on a single source with limited data, or their performance has been unsatisfactory. To fill this gap, we propose a method called UNISELF, which achieves high accuracy within a single training domain while demonstrating strong generalizability across multiple out-of-domain test datasets. UNISELF employs a novel test-time self-ensembled lesion fusion to improve segmentation accuracy, and leverages test-time instance normalization (TTIN) of latent features to address domain shifts and missing input contrasts. Trained on the ISBI 2015 longitudinal MS segmentation challenge training dataset, UNISELF ranks among the best-performing methods on the challenge test dataset. Additionally, UNISELF outperforms all benchmark methods trained on the same ISBI training data across diverse out-of-domain test datasets with domain shifts and missing contrasts, including the public MICCAI 2016 and UMCL datasets, as well as a private multisite dataset. These test datasets exhibit domain shifts and/or missing contrasts caused by variations in acquisition protocols, scanner types, and imaging artifacts arising from imperfect acquisition. Our code is available at https://github.com/uponacceptance.
Brightness-Invariant Tracking Estimation in Tagged MRI
May 23, 2025Abstract:Magnetic resonance (MR) tagging is an imaging technique for noninvasively tracking tissue motion in vivo by creating a visible pattern of magnetization saturation (tags) that deforms with the tissue. Due to longitudinal relaxation and progression to steady-state, the tags and tissue brightnesses change over time, which makes tracking with optical flow methods error-prone. Although Fourier methods can alleviate these problems, they are also sensitive to brightness changes as well as spectral spreading due to motion. To address these problems, we introduce the brightness-invariant tracking estimation (BRITE) technique for tagged MRI. BRITE disentangles the anatomy from the tag pattern in the observed tagged image sequence and simultaneously estimates the Lagrangian motion. The inherent ill-posedness of this problem is addressed by leveraging the expressive power of denoising diffusion probabilistic models to represent the probabilistic distribution of the underlying anatomy and the flexibility of physics-informed neural networks to estimate biologically-plausible motion. A set of tagged MR images of a gel phantom was acquired with various tag periods and imaging flip angles to demonstrate the impact of brightness variations and to validate our method. The results show that BRITE achieves more accurate motion and strain estimates as compared to other state of the art methods, while also being resistant to tag fading.
ECLARE: Efficient cross-planar learning for anisotropic resolution enhancement
Mar 14, 2025Abstract:In clinical imaging, magnetic resonance (MR) image volumes are often acquired as stacks of 2D slices, permitting decreased scan times, improved signal-to-noise ratio, and image contrasts unique to 2D MR pulse sequences. While this is sufficient for clinical evaluation, automated algorithms designed for 3D analysis perform sub-optimally on 2D-acquired scans, especially those with thick slices and gaps between slices. Super-resolution (SR) methods aim to address this problem, but previous methods do not address all of the following: slice profile shape estimation, slice gap, domain shift, and non-integer / arbitrary upsampling factors. In this paper, we propose ECLARE (Efficient Cross-planar Learning for Anisotropic Resolution Enhancement), a self-SR method that addresses each of these factors. ECLARE estimates the slice profile from the 2D-acquired multi-slice MR volume, trains a network to learn the mapping from low-resolution to high-resolution in-plane patches from the same volume, and performs SR with anti-aliasing. We compared ECLARE to cubic B-spline interpolation, SMORE, and other contemporary SR methods. We used realistic and representative simulations so that quantitative performance against a ground truth could be computed, and ECLARE outperformed all other methods in both signal recovery and downstream tasks. On real data for which there is no ground truth, ECLARE demonstrated qualitative superiority over other methods as well. Importantly, as ECLARE does not use external training data it cannot suffer from domain shift between training and testing. Our code is open-source and available at https://www.github.com/sremedios/eclare.
Unique MS Lesion Identification from MRI
Oct 12, 2024



Abstract:Unique identification of multiple sclerosis (MS) white matter lesions (WMLs) is important to help characterize MS progression. WMLs are routinely identified from magnetic resonance images (MRIs) but the resultant total lesion load does not correlate well with EDSS; whereas mean unique lesion volume has been shown to correlate with EDSS. Our approach builds on prior work by incorporating Hessian matrix computation from lesion probability maps before using the random walker algorithm to estimate the volume of each unique lesion. Synthetic images demonstrate our ability to accurately count the number of lesions present. The takeaways, are: 1) that our method correctly identifies all lesions including many that are missed by previous methods; 2) we can better separate confluent lesions; and 3) we can accurately capture the total volume of WMLs in a given probability map. This work will allow new more meaningful statistics to be computed from WMLs in brain MRIs
Influence of Early through Late Fusion on Pancreas Segmentation from Imperfectly Registered Multimodal MRI
Sep 06, 2024Abstract:Multimodal fusion promises better pancreas segmentation. However, where to perform fusion in models is still an open question. It is unclear if there is a best location to fuse information when analyzing pairs of imperfectly aligned images. Two main alignment challenges in this pancreas segmentation study are 1) the pancreas is deformable and 2) breathing deforms the abdomen. Even after image registration, relevant deformations are often not corrected. We examine how early through late fusion impacts pancreas segmentation. We used 353 pairs of T2-weighted (T2w) and T1-weighted (T1w) abdominal MR images from 163 subjects with accompanying pancreas labels. We used image registration (deeds) to align the image pairs. We trained a collection of basic UNets with different fusion points, spanning from early to late, to assess how early through late fusion influenced segmentation performance on imperfectly aligned images. We assessed generalization of fusion points on nnUNet. The single-modality T2w baseline using a basic UNet model had a Dice score of 0.73, while the same baseline on the nnUNet model achieved 0.80. For the basic UNet, the best fusion approach occurred in the middle of the encoder (early/mid fusion), which led to a statistically significant improvement of 0.0125 on Dice score compared to the baseline. For the nnUNet, the best fusion approach was na\"ive image concatenation before the model (early fusion), which resulted in a statistically significant Dice score increase of 0.0021 compared to baseline. Fusion in specific blocks can improve performance, but the best blocks for fusion are model specific, and the gains are small. In imperfectly registered datasets, fusion is a nuanced problem, with the art of design remaining vital for uncovering potential insights. Future innovation is needed to better address fusion in cases of imperfect alignment of abdominal image pairs.
Beyond MR Image Harmonization: Resolution Matters Too
Aug 30, 2024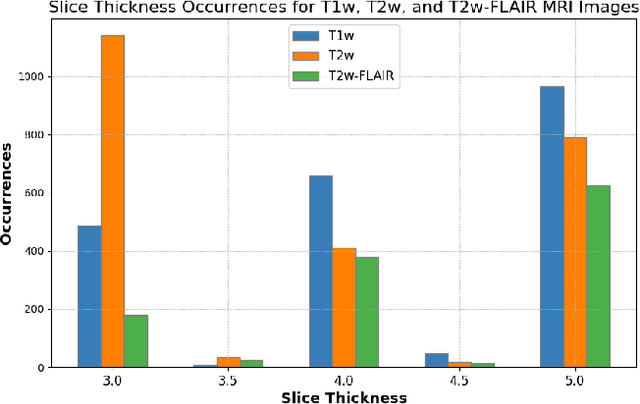
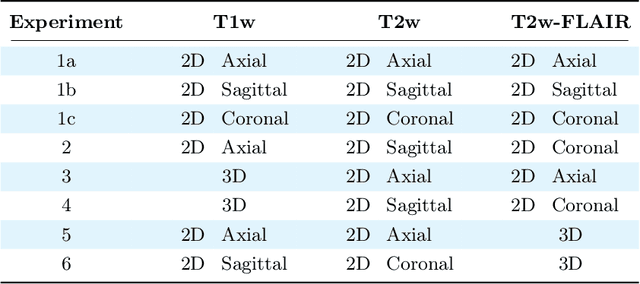
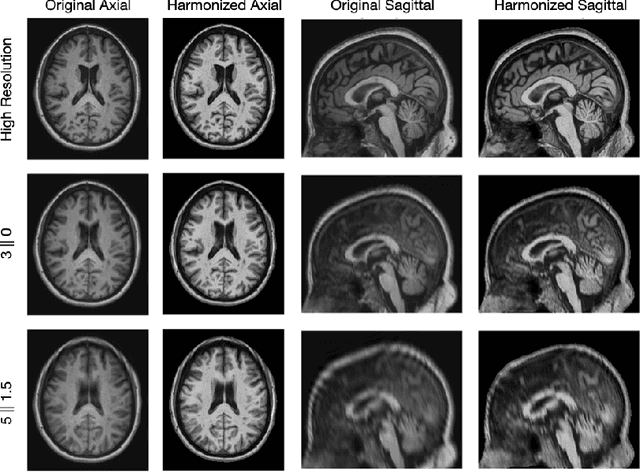
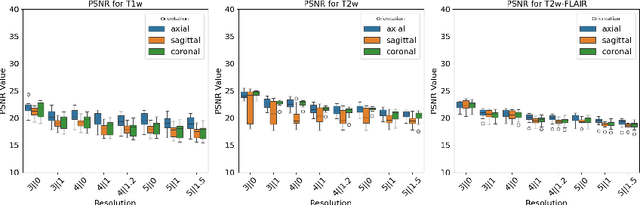
Abstract:Magnetic resonance (MR) imaging is commonly used in the clinical setting to non-invasively monitor the body. There exists a large variability in MR imaging due to differences in scanner hardware, software, and protocol design. Ideally, a processing algorithm should perform robustly to this variability, but that is not always the case in reality. This introduces a need for image harmonization to overcome issues of domain shift when performing downstream analysis such as segmentation. Most image harmonization models focus on acquisition parameters such as inversion time or repetition time, but they ignore an important aspect in MR imaging -- resolution. In this paper, we evaluate the impact of image resolution on harmonization using a pretrained harmonization algorithm. We simulate 2D acquisitions of various slice thicknesses and gaps from 3D acquired, 1mm3 isotropic MR images and demonstrate how the performance of a state-of-the-art image harmonization algorithm varies as resolution changes. We discuss the most ideal scenarios for image resolution including acquisition orientation when 3D imaging is not available, which is common for many clinical scanners. Our results show that harmonization on low-resolution images does not account for acquisition resolution and orientation variations. Super-resolution can be used to alleviate resolution variations but it is not always used. Our methodology can generalize to help evaluate the impact of image acquisition resolution for multiple tasks. Determining the limits of a pretrained algorithm is important when considering preprocessing steps and trust in the results.
Nucleus subtype classification using inter-modality learning
Jan 29, 2024Abstract:Understanding the way cells communicate, co-locate, and interrelate is essential to understanding human physiology. Hematoxylin and eosin (H&E) staining is ubiquitously available both for clinical studies and research. The Colon Nucleus Identification and Classification (CoNIC) Challenge has recently innovated on robust artificial intelligence labeling of six cell types on H&E stains of the colon. However, this is a very small fraction of the number of potential cell classification types. Specifically, the CoNIC Challenge is unable to classify epithelial subtypes (progenitor, endocrine, goblet), lymphocyte subtypes (B, helper T, cytotoxic T), or connective subtypes (fibroblasts, stromal). In this paper, we propose to use inter-modality learning to label previously un-labelable cell types on virtual H&E. We leveraged multiplexed immunofluorescence (MxIF) histology imaging to identify 14 subclasses of cell types. We performed style transfer to synthesize virtual H&E from MxIF and transferred the higher density labels from MxIF to these virtual H&E images. We then evaluated the efficacy of learning in this approach. We identified helper T and progenitor nuclei with positive predictive values of $0.34 \pm 0.15$ (prevalence $0.03 \pm 0.01$) and $0.47 \pm 0.1$ (prevalence $0.07 \pm 0.02$) respectively on virtual H&E. This approach represents a promising step towards automating annotation in digital pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge