Ruining Deng
Patient-Conditioned Adaptive Offsets for Reliable Diagnosis across Subgroups
Jan 19, 2026Abstract:AI models for medical diagnosis often exhibit uneven performance across patient populations due to heterogeneity in disease prevalence, imaging appearance, and clinical risk profiles. Existing algorithmic fairness approaches typically seek to reduce such disparities by suppressing sensitive attributes. However, in medical settings these attributes often carry essential diagnostic information, and removing them can degrade accuracy and reliability, particularly in high-stakes applications. In contrast, clinical decision making explicitly incorporates patient context when interpreting diagnostic evidence, suggesting a different design direction for subgroup-aware models. In this paper, we introduce HyperAdapt, a patient-conditioned adaptation framework that improves subgroup reliability while maintaining a shared diagnostic model. Clinically relevant attributes such as age and sex are encoded into a compact embedding and used to condition a hypernetwork-style module, which generates small residual modulation parameters for selected layers of a shared backbone. This design preserves the general medical knowledge learned by the backbone while enabling targeted adjustments that reflect patient-specific variability. To ensure efficiency and robustness, adaptations are constrained through low-rank and bottlenecked parameterizations, limiting both model complexity and computational overhead. Experiments across multiple public medical imaging benchmarks demonstrate that the proposed approach consistently improves subgroup-level performance without sacrificing overall accuracy. On the PAD-UFES-20 dataset, our method outperforms the strongest competing baseline by 4.1% in recall and 4.4% in F1 score, with larger gains observed for underrepresented patient populations.
SCR2-ST: Combine Single Cell with Spatial Transcriptomics for Efficient Active Sampling via Reinforcement Learning
Dec 15, 2025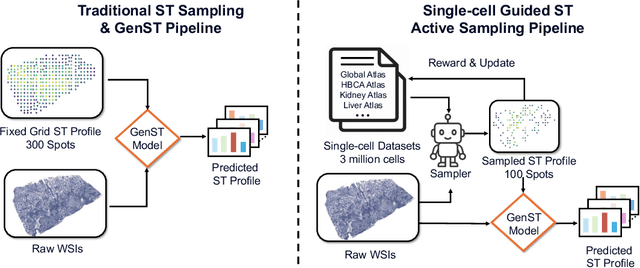
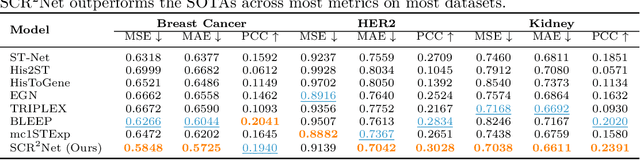
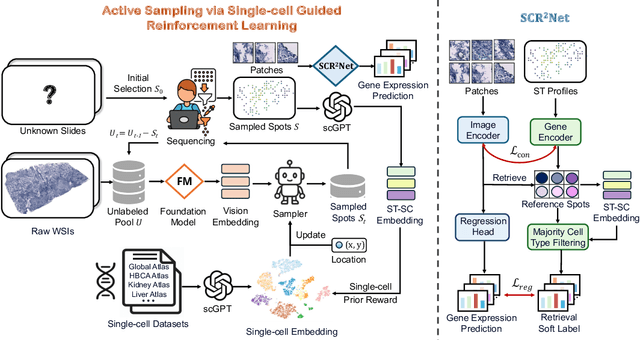
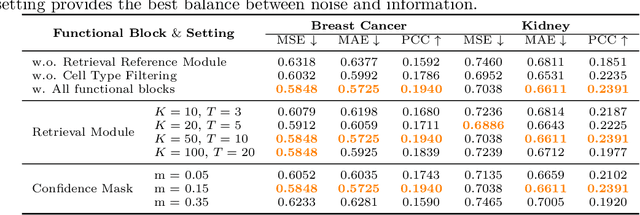
Abstract:Spatial transcriptomics (ST) is an emerging technology that enables researchers to investigate the molecular relationships underlying tissue morphology. However, acquiring ST data remains prohibitively expensive, and traditional fixed-grid sampling strategies lead to redundant measurements of morphologically similar or biologically uninformative regions, thus resulting in scarce data that constrain current methods. The well-established single-cell sequencing field, however, could provide rich biological data as an effective auxiliary source to mitigate this limitation. To bridge these gaps, we introduce SCR2-ST, a unified framework that leverages single-cell prior knowledge to guide efficient data acquisition and accurate expression prediction. SCR2-ST integrates a single-cell guided reinforcement learning-based (SCRL) active sampling and a hybrid regression-retrieval prediction network SCR2Net. SCRL combines single-cell foundation model embeddings with spatial density information to construct biologically grounded reward signals, enabling selective acquisition of informative tissue regions under constrained sequencing budgets. SCR2Net then leverages the actively sampled data through a hybrid architecture combining regression-based modeling with retrieval-augmented inference, where a majority cell-type filtering mechanism suppresses noisy matches and retrieved expression profiles serve as soft labels for auxiliary supervision. We evaluated SCR2-ST on three public ST datasets, demonstrating SOTA performance in both sampling efficiency and prediction accuracy, particularly under low-budget scenarios. Code is publicly available at: https://github.com/hrlblab/SCR2ST
From Classification to Cross-Modal Understanding: Leveraging Vision-Language Models for Fine-Grained Renal Pathology
Nov 15, 2025Abstract:Fine-grained glomerular subtyping is central to kidney biopsy interpretation, but clinically valuable labels are scarce and difficult to obtain. Existing computational pathology approaches instead tend to evaluate coarse diseased classification under full supervision with image-only models, so it remains unclear how vision-language models (VLMs) should be adapted for clinically meaningful subtyping under data constraints. In this work, we model fine-grained glomerular subtyping as a clinically realistic few-shot problem and systematically evaluate both pathology-specialized and general-purpose vision-language models under this setting. We assess not only classification performance (accuracy, AUC, F1) but also the geometry of the learned representations, examining feature alignment between image and text embeddings and the separability of glomerular subtypes. By jointly analyzing shot count, model architecture and domain knowledge, and adaptation strategy, this study provides guidance for future model selection and training under real clinical data constraints. Our results indicate that pathology-specialized vision-language backbones, when paired with the vanilla fine-tuning, are the most effective starting point. Even with only 4-8 labeled examples per glomeruli subtype, these models begin to capture distinctions and show substantial gains in discrimination and calibration, though additional supervision continues to yield incremental improvements. We also find that the discrimination between positive and negative examples is as important as image-text alignment. Overall, our results show that supervision level and adaptation strategy jointly shape both diagnostic performance and multimodal structure, providing guidance for model selection, adaptation strategies, and annotation investment.
How Close Are We? Limitations and Progress of AI Models in Banff Lesion Scoring
Oct 31, 2025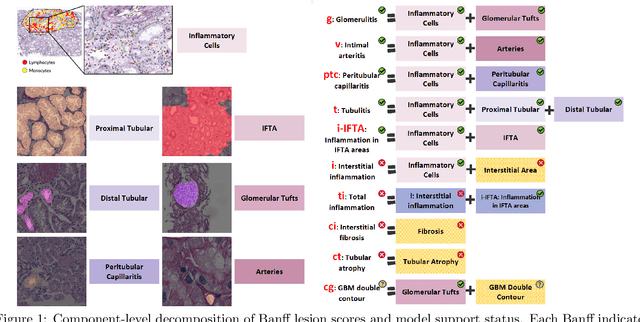
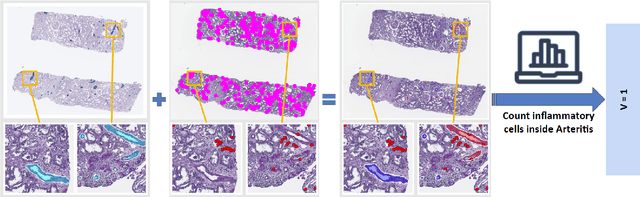
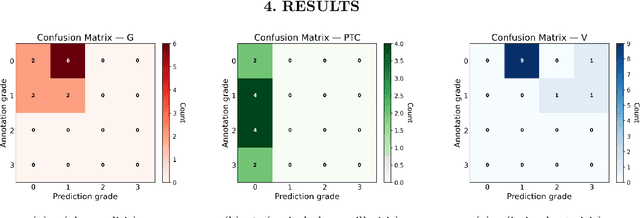
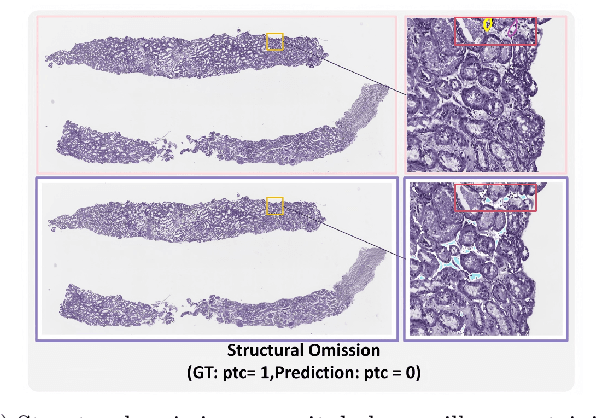
Abstract:The Banff Classification provides the global standard for evaluating renal transplant biopsies, yet its semi-quantitative nature, complex criteria, and inter-observer variability present significant challenges for computational replication. In this study, we explore the feasibility of approximating Banff lesion scores using existing deep learning models through a modular, rule-based framework. We decompose each Banff indicator - such as glomerulitis (g), peritubular capillaritis (ptc), and intimal arteritis (v) - into its constituent structural and inflammatory components, and assess whether current segmentation and detection tools can support their computation. Model outputs are mapped to Banff scores using heuristic rules aligned with expert guidelines, and evaluated against expert-annotated ground truths. Our findings highlight both partial successes and critical failure modes, including structural omission, hallucination, and detection ambiguity. Even when final scores match expert annotations, inconsistencies in intermediate representations often undermine interpretability. These results reveal the limitations of current AI pipelines in replicating computational expert-level grading, and emphasize the importance of modular evaluation and computational Banff grading standard in guiding future model development for transplant pathology.
M^3-GloDets: Multi-Region and Multi-Scale Analysis of Fine-Grained Diseased Glomerular Detection
Aug 25, 2025Abstract:Accurate detection of diseased glomeruli is fundamental to progress in renal pathology and underpins the delivery of reliable clinical diagnoses. Although recent advances in computer vision have produced increasingly sophisticated detection algorithms, the majority of research efforts have focused on normal glomeruli or instances of global sclerosis, leaving the wider spectrum of diseased glomerular subtypes comparatively understudied. This disparity is not without consequence; the nuanced and highly variable morphological characteristics that define these disease variants frequently elude even the most advanced computational models. Moreover, ongoing debate surrounds the choice of optimal imaging magnifications and region-of-view dimensions for fine-grained glomerular analysis, adding further complexity to the pursuit of accurate classification and robust segmentation. To bridge these gaps, we present M^3-GloDet, a systematic framework designed to enable thorough evaluation of detection models across a broad continuum of regions, scales, and classes. Within this framework, we evaluate both long-standing benchmark architectures and recently introduced state-of-the-art models that have achieved notable performance, using an experimental design that reflects the diversity of region-of-interest sizes and imaging resolutions encountered in routine digital renal pathology. As the results, we found that intermediate patch sizes offered the best balance between context and efficiency. Additionally, moderate magnifications enhanced generalization by reducing overfitting. Through systematic comparison of these approaches on a multi-class diseased glomerular dataset, our aim is to advance the understanding of model strengths and limitations, and to offer actionable insights for the refinement of automated detection strategies and clinical workflows in the digital pathology domain.
DyMorph-B2I: Dynamic and Morphology-Guided Binary-to-Instance Segmentation for Renal Pathology
Aug 21, 2025Abstract:Accurate morphological quantification of renal pathology functional units relies on instance-level segmentation, yet most existing datasets and automated methods provide only binary (semantic) masks, limiting the precision of downstream analyses. Although classical post-processing techniques such as watershed, morphological operations, and skeletonization, are often used to separate semantic masks into instances, their individual effectiveness is constrained by the diverse morphologies and complex connectivity found in renal tissue. In this study, we present DyMorph-B2I, a dynamic, morphology-guided binary-to-instance segmentation pipeline tailored for renal pathology. Our approach integrates watershed, skeletonization, and morphological operations within a unified framework, complemented by adaptive geometric refinement and customizable hyperparameter tuning for each class of functional unit. Through systematic parameter optimization, DyMorph-B2I robustly separates adherent and heterogeneous structures present in binary masks. Experimental results demonstrate that our method outperforms individual classical approaches and na\"ive combinations, enabling superior instance separation and facilitating more accurate morphometric analysis in renal pathology workflows. The pipeline is publicly available at: https://github.com/ddrrnn123/DyMorph-B2I.
Img2ST-Net: Efficient High-Resolution Spatial Omics Prediction from Whole Slide Histology Images via Fully Convolutional Image-to-Image Learning
Aug 20, 2025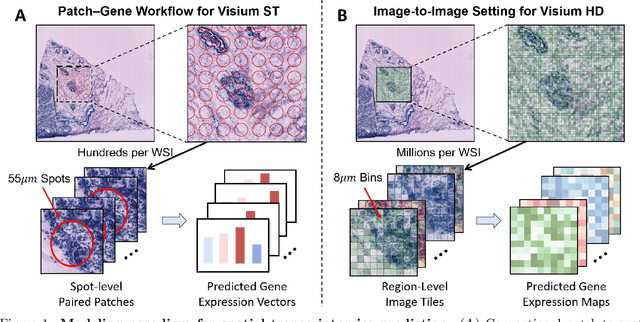
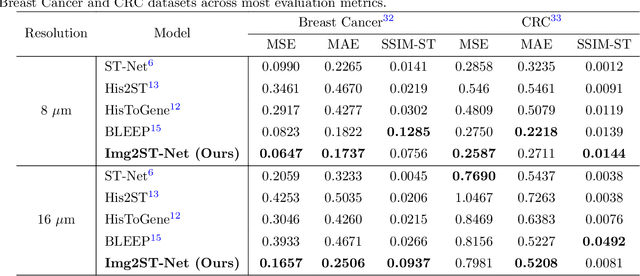
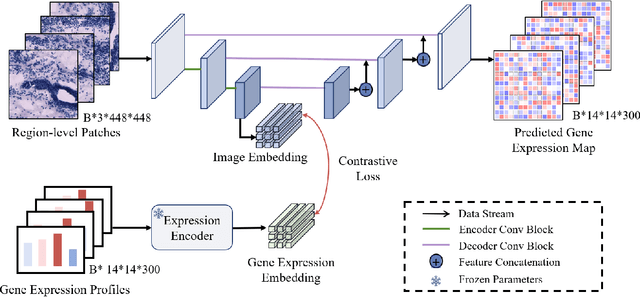
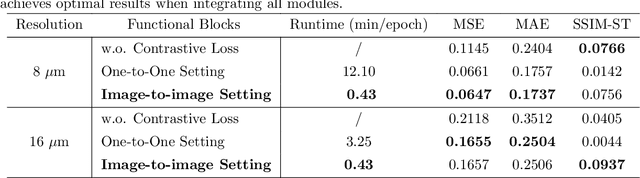
Abstract:Recent advances in multi-modal AI have demonstrated promising potential for generating the currently expensive spatial transcriptomics (ST) data directly from routine histology images, offering a means to reduce the high cost and time-intensive nature of ST data acquisition. However, the increasing resolution of ST, particularly with platforms such as Visium HD achieving 8um or finer, introduces significant computational and modeling challenges. Conventional spot-by-spot sequential regression frameworks become inefficient and unstable at this scale, while the inherent extreme sparsity and low expression levels of high-resolution ST further complicate both prediction and evaluation. To address these limitations, we propose Img2ST-Net, a novel histology-to-ST generation framework for efficient and parallel high-resolution ST prediction. Unlike conventional spot-by-spot inference methods, Img2ST-Net employs a fully convolutional architecture to generate dense, HD gene expression maps in a parallelized manner. By modeling HD ST data as super-pixel representations, the task is reformulated from image-to-omics inference into a super-content image generation problem with hundreds or thousands of output channels. This design not only improves computational efficiency but also better preserves the spatial organization intrinsic to spatial omics data. To enhance robustness under sparse expression patterns, we further introduce SSIM-ST, a structural-similarity-based evaluation metric tailored for high-resolution ST analysis. We present a scalable, biologically coherent framework for high-resolution ST prediction. Img2ST-Net offers a principled solution for efficient and accurate ST inference at scale. Our contributions lay the groundwork for next-generation ST modeling that is robust and resolution-aware. The source code has been made publicly available at https://github.com/hrlblab/Img2ST-Net.
Quantitative Benchmarking of Anomaly Detection Methods in Digital Pathology
Jun 24, 2025Abstract:Anomaly detection has been widely studied in the context of industrial defect inspection, with numerous methods developed to tackle a range of challenges. In digital pathology, anomaly detection holds significant potential for applications such as rare disease identification, artifact detection, and biomarker discovery. However, the unique characteristics of pathology images, such as their large size, multi-scale structures, stain variability, and repetitive patterns, introduce new challenges that current anomaly detection algorithms struggle to address. In this quantitative study, we benchmark over 20 classical and prevalent anomaly detection methods through extensive experiments. We curated five digital pathology datasets, both real and synthetic, to systematically evaluate these approaches. Our experiments investigate the influence of image scale, anomaly pattern types, and training epoch selection strategies on detection performance. The results provide a detailed comparison of each method's strengths and limitations, establishing a comprehensive benchmark to guide future research in anomaly detection for digital pathology images.
IRS: Incremental Relationship-guided Segmentation for Digital Pathology
May 28, 2025Abstract:Continual learning is rapidly emerging as a key focus in computer vision, aiming to develop AI systems capable of continuous improvement, thereby enhancing their value and practicality in diverse real-world applications. In healthcare, continual learning holds great promise for continuously acquired digital pathology data, which is collected in hospitals on a daily basis. However, panoramic segmentation on digital whole slide images (WSIs) presents significant challenges, as it is often infeasible to obtain comprehensive annotations for all potential objects, spanning from coarse structures (e.g., regions and unit objects) to fine structures (e.g., cells). This results in temporally and partially annotated data, posing a major challenge in developing a holistic segmentation framework. Moreover, an ideal segmentation model should incorporate new phenotypes, unseen diseases, and diverse populations, making this task even more complex. In this paper, we introduce a novel and unified Incremental Relationship-guided Segmentation (IRS) learning scheme to address temporally acquired, partially annotated data while maintaining out-of-distribution (OOD) continual learning capacity in digital pathology. The key innovation of IRS lies in its ability to realize a new spatial-temporal OOD continual learning paradigm by mathematically modeling anatomical relationships between existing and newly introduced classes through a simple incremental universal proposition matrix. Experimental results demonstrate that the IRS method effectively handles the multi-scale nature of pathological segmentation, enabling precise kidney segmentation across various structures (regions, units, and cells) as well as OOD disease lesions at multiple magnifications. This capability significantly enhances domain generalization, making IRS a robust approach for real-world digital pathology applications.
DeepAndes: A Self-Supervised Vision Foundation Model for Multi-Spectral Remote Sensing Imagery of the Andes
Apr 28, 2025Abstract:By mapping sites at large scales using remotely sensed data, archaeologists can generate unique insights into long-term demographic trends, inter-regional social networks, and past adaptations to climate change. Remote sensing surveys complement field-based approaches, and their reach can be especially great when combined with deep learning and computer vision techniques. However, conventional supervised deep learning methods face challenges in annotating fine-grained archaeological features at scale. While recent vision foundation models have shown remarkable success in learning large-scale remote sensing data with minimal annotations, most off-the-shelf solutions are designed for RGB images rather than multi-spectral satellite imagery, such as the 8-band data used in our study. In this paper, we introduce DeepAndes, a transformer-based vision foundation model trained on three million multi-spectral satellite images, specifically tailored for Andean archaeology. DeepAndes incorporates a customized DINOv2 self-supervised learning algorithm optimized for 8-band multi-spectral imagery, marking the first foundation model designed explicitly for the Andes region. We evaluate its image understanding performance through imbalanced image classification, image instance retrieval, and pixel-level semantic segmentation tasks. Our experiments show that DeepAndes achieves superior F1 scores, mean average precision, and Dice scores in few-shot learning scenarios, significantly outperforming models trained from scratch or pre-trained on smaller datasets. This underscores the effectiveness of large-scale self-supervised pre-training in archaeological remote sensing. Codes will be available on https://github.com/geopacha/DeepAndes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge