Daguang Xu
Humanoid Robots as First Assistants in Endoscopic Surgery
Feb 27, 2026Abstract:Humanoid robots have become a focal point of technological ambition, with claims of surgical capability within years in mainstream discourse. These projections are aspirational yet lack empirical grounding. To date, no humanoid has assisted a surgeon through an actual procedure, let alone performed one. The work described here breaks this new ground. Here we report a proof of concept in which a teleoperated Unitree G1 provided endoscopic visualization while an attending otolaryngologist performed a cadaveric sphenoidectomy. The procedure was completed successfully, with stable visualization maintained throughout. Teleoperation allowed assessment of whether the humanoid form factor could meet the physical demands of surgical assistance in terms of sustenance and precision; the cognitive demands were satisfied -- for now -- by the operator. Post-procedure analysis identified engineering targets for clinical translation, alongside near-term opportunities such as autonomous diagnostic scoping. This work establishes form-factor feasibility for humanoid surgical assistance while identifying challenges for continued development.
LUMEN: Longitudinal Multi-Modal Radiology Model for Prognosis and Diagnosis
Feb 24, 2026Abstract:Large vision-language models (VLMs) have evolved from general-purpose applications to specialized use cases such as in the clinical domain, demonstrating potential for decision support in radiology. One promising application is assisting radiologists in decision-making by the analysis of radiology imaging data such as chest X-rays (CXR) via a visual and natural language question-answering (VQA) interface. When longitudinal imaging is available, radiologists analyze temporal changes, which are essential for accurate diagnosis and prognosis. The manual longitudinal analysis is a time-consuming process, motivating the development of a training framework that can provide prognostic capabilities. We introduce a novel training framework LUMEN, that is optimized for longitudinal CXR interpretation, leveraging multi-image and multi-task instruction fine-tuning to enhance prognostic and diagnostic performance. We conduct experiments on the publicly available MIMIC-CXR and its associated Medical-Diff-VQA datasets. We further formulate and construct a novel instruction-following dataset incorporating longitudinal studies, enabling the development of a prognostic VQA task. Our method demonstrates significant improvements over baseline models in diagnostic VQA tasks, and more importantly, shows promising potential for prognostic capabilities. These results underscore the value of well-designed, instruction-tuned VLMs in enabling more accurate and clinically meaningful radiological interpretation of longitudinal radiological imaging data.
VISTA-PATH: An interactive foundation model for pathology image segmentation and quantitative analysis in computational pathology
Jan 23, 2026Abstract:Accurate semantic segmentation for histopathology image is crucial for quantitative tissue analysis and downstream clinical modeling. Recent segmentation foundation models have improved generalization through large-scale pretraining, yet remain poorly aligned with pathology because they treat segmentation as a static visual prediction task. Here we present VISTA-PATH, an interactive, class-aware pathology segmentation foundation model designed to resolve heterogeneous structures, incorporate expert feedback, and produce pixel-level segmentation that are directly meaningful for clinical interpretation. VISTA-PATH jointly conditions segmentation on visual context, semantic tissue descriptions, and optional expert-provided spatial prompts, enabling precise multi-class segmentation across heterogeneous pathology images. To support this paradigm, we curate VISTA-PATH Data, a large-scale pathology segmentation corpus comprising over 1.6 million image-mask-text triplets spanning 9 organs and 93 tissue classes. Across extensive held-out and external benchmarks, VISTA-PATH consistently outperforms existing segmentation foundation models. Importantly, VISTA-PATH supports dynamic human-in-the-loop refinement by propagating sparse, patch-level bounding-box annotation feedback into whole-slide segmentation. Finally, we show that the high-fidelity, class-aware segmentation produced by VISTA-PATH is a preferred model for computational pathology. It improve tissue microenvironment analysis through proposed Tumor Interaction Score (TIS), which exhibits strong and significant associations with patient survival. Together, these results establish VISTA-PATH as a foundation model that elevates pathology image segmentation from a static prediction to an interactive and clinically grounded representation for digital pathology. Source code and demo can be found at https://github.com/zhihuanglab/VISTA-PATH.
SurgWorld: Learning Surgical Robot Policies from Videos via World Modeling
Dec 30, 2025Abstract:Data scarcity remains a fundamental barrier to achieving fully autonomous surgical robots. While large scale vision language action (VLA) models have shown impressive generalization in household and industrial manipulation by leveraging paired video action data from diverse domains, surgical robotics suffers from the paucity of datasets that include both visual observations and accurate robot kinematics. In contrast, vast corpora of surgical videos exist, but they lack corresponding action labels, preventing direct application of imitation learning or VLA training. In this work, we aim to alleviate this problem by learning policy models from SurgWorld, a world model designed for surgical physical AI. We curated the Surgical Action Text Alignment (SATA) dataset with detailed action description specifically for surgical robots. Then we built SurgeWorld based on the most advanced physical AI world model and SATA. It's able to generate diverse, generalizable and realistic surgery videos. We are also the first to use an inverse dynamics model to infer pseudokinematics from synthetic surgical videos, producing synthetic paired video action data. We demonstrate that a surgical VLA policy trained with these augmented data significantly outperforms models trained only on real demonstrations on a real surgical robot platform. Our approach offers a scalable path toward autonomous surgical skill acquisition by leveraging the abundance of unlabeled surgical video and generative world modeling, thus opening the door to generalizable and data efficient surgical robot policies.
SDUM: A Scalable Deep Unrolled Model for Universal MRI Reconstruction
Dec 19, 2025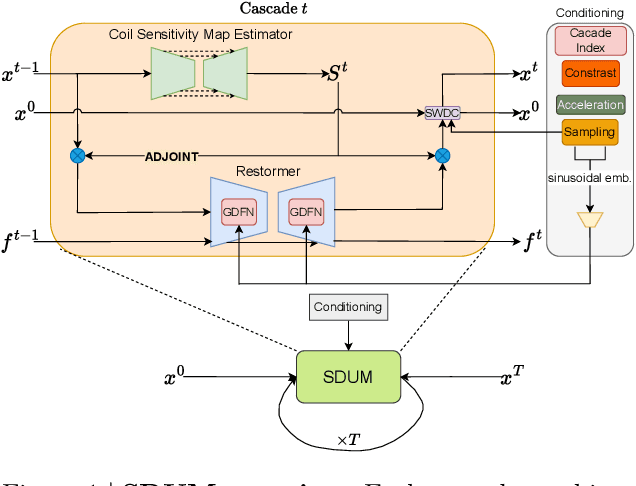

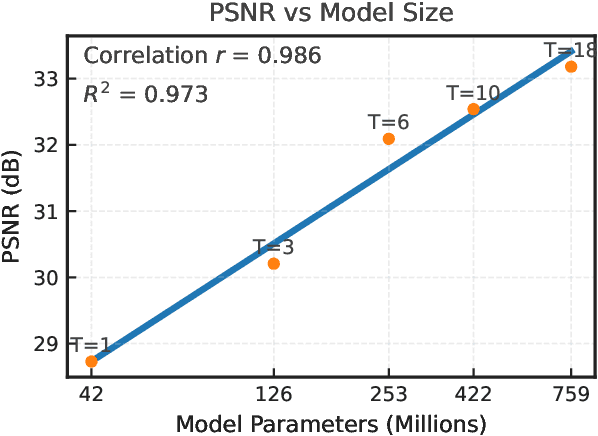
Abstract:Clinical MRI encompasses diverse imaging protocols--spanning anatomical targets (cardiac, brain, knee), contrasts (T1, T2, mapping), sampling patterns (Cartesian, radial, spiral, kt-space), and acceleration factors--yet current deep learning reconstructions are typically protocol-specific, hindering generalization and deployment. We introduce Scalable Deep Unrolled Model (SDUM), a universal framework combining a Restormer-based reconstructor, a learned coil sensitivity map estimator (CSME), sampling-aware weighted data consistency (SWDC), universal conditioning (UC) on cascade index and protocol metadata, and progressive cascade expansion training. SDUM exhibits foundation-model-like scaling behavior: reconstruction quality follows PSNR ${\sim}$ log(parameters) with correlation $r{=}0.986$ ($R^2{=}0.973$) up to 18 cascades, demonstrating predictable performance gains with model depth. A single SDUM trained on heterogeneous data achieves state-of-the-art results across all four CMRxRecon2025 challenge tracks--multi-center, multi-disease, 5T, and pediatric--without task-specific fine-tuning, surpassing specialized baselines by up to ${+}1.0$~dB. On CMRxRecon2024, SDUM outperforms the winning method PromptMR+ by ${+}0.55$~dB; on fastMRI brain, it exceeds PC-RNN by ${+}1.8$~dB. Ablations validate each component: SWDC ${+}0.43$~dB over standard DC, per-cascade CSME ${+}0.51$~dB, UC ${+}0.38$~dB. These results establish SDUM as a practical path toward universal, scalable MRI reconstruction.
See More, Change Less: Anatomy-Aware Diffusion for Contrast Enhancement
Dec 08, 2025Abstract:Image enhancement improves visual quality and helps reveal details that are hard to see in the original image. In medical imaging, it can support clinical decision-making, but current models often over-edit. This can distort organs, create false findings, and miss small tumors because these models do not understand anatomy or contrast dynamics. We propose SMILE, an anatomy-aware diffusion model that learns how organs are shaped and how they take up contrast. It enhances only clinically relevant regions while leaving all other areas unchanged. SMILE introduces three key ideas: (1) structure-aware supervision that follows true organ boundaries and contrast patterns; (2) registration-free learning that works directly with unaligned multi-phase CT scans; (3) unified inference that provides fast and consistent enhancement across all contrast phases. Across six external datasets, SMILE outperforms existing methods in image quality (14.2% higher SSIM, 20.6% higher PSNR, 50% better FID) and in clinical usefulness by producing anatomically accurate and diagnostically meaningful images. SMILE also improves cancer detection from non-contrast CT, raising the F1 score by up to 10 percent.
Better Tokens for Better 3D: Advancing Vision-Language Modeling in 3D Medical Imaging
Oct 23, 2025Abstract:Recent progress in vision-language modeling for 3D medical imaging has been fueled by large-scale computed tomography (CT) corpora with paired free-text reports, stronger architectures, and powerful pretrained models. This has enabled applications such as automated report generation and text-conditioned 3D image synthesis. Yet, current approaches struggle with high-resolution, long-sequence volumes: contrastive pretraining often yields vision encoders that are misaligned with clinical language, and slice-wise tokenization blurs fine anatomy, reducing diagnostic performance on downstream tasks. We introduce BTB3D (Better Tokens for Better 3D), a causal convolutional encoder-decoder that unifies 2D and 3D training and inference while producing compact, frequency-aware volumetric tokens. A three-stage training curriculum enables (i) local reconstruction, (ii) overlapping-window tiling, and (iii) long-context decoder refinement, during which the model learns from short slice excerpts yet generalizes to scans exceeding 300 slices without additional memory overhead. BTB3D sets a new state-of-the-art on two key tasks: it improves BLEU scores and increases clinical F1 by 40% over CT2Rep, CT-CHAT, and Merlin for report generation; and it reduces FID by 75% and halves FVD compared to GenerateCT and MedSyn for text-to-CT synthesis, producing anatomically consistent 512*512*241 volumes. These results confirm that precise three-dimensional tokenization, rather than larger language backbones alone, is essential for scalable vision-language modeling in 3D medical imaging. The codebase is available at: https://github.com/ibrahimethemhamamci/BTB3D
Img2ST-Net: Efficient High-Resolution Spatial Omics Prediction from Whole Slide Histology Images via Fully Convolutional Image-to-Image Learning
Aug 20, 2025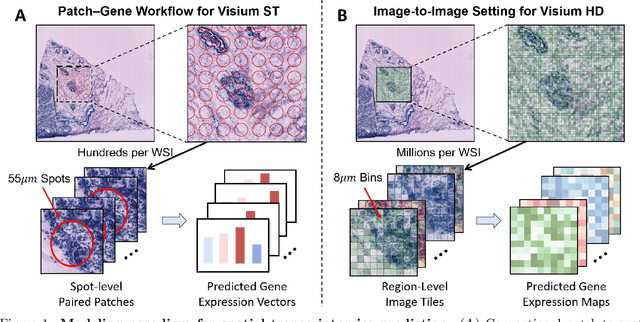
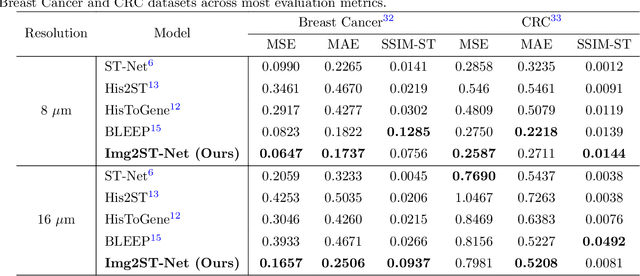
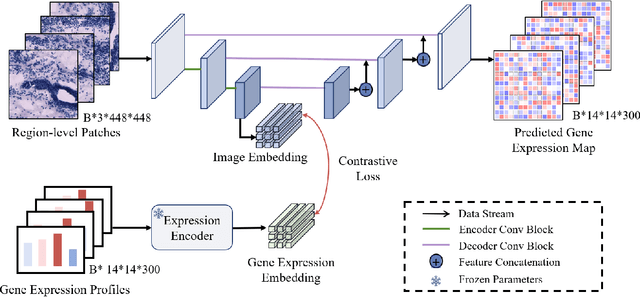
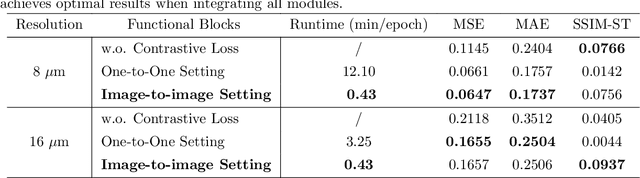
Abstract:Recent advances in multi-modal AI have demonstrated promising potential for generating the currently expensive spatial transcriptomics (ST) data directly from routine histology images, offering a means to reduce the high cost and time-intensive nature of ST data acquisition. However, the increasing resolution of ST, particularly with platforms such as Visium HD achieving 8um or finer, introduces significant computational and modeling challenges. Conventional spot-by-spot sequential regression frameworks become inefficient and unstable at this scale, while the inherent extreme sparsity and low expression levels of high-resolution ST further complicate both prediction and evaluation. To address these limitations, we propose Img2ST-Net, a novel histology-to-ST generation framework for efficient and parallel high-resolution ST prediction. Unlike conventional spot-by-spot inference methods, Img2ST-Net employs a fully convolutional architecture to generate dense, HD gene expression maps in a parallelized manner. By modeling HD ST data as super-pixel representations, the task is reformulated from image-to-omics inference into a super-content image generation problem with hundreds or thousands of output channels. This design not only improves computational efficiency but also better preserves the spatial organization intrinsic to spatial omics data. To enhance robustness under sparse expression patterns, we further introduce SSIM-ST, a structural-similarity-based evaluation metric tailored for high-resolution ST analysis. We present a scalable, biologically coherent framework for high-resolution ST prediction. Img2ST-Net offers a principled solution for efficient and accurate ST inference at scale. Our contributions lay the groundwork for next-generation ST modeling that is robust and resolution-aware. The source code has been made publicly available at https://github.com/hrlblab/Img2ST-Net.
PanTS: The Pancreatic Tumor Segmentation Dataset
Jul 02, 2025Abstract:PanTS is a large-scale, multi-institutional dataset curated to advance research in pancreatic CT analysis. It contains 36,390 CT scans from 145 medical centers, with expert-validated, voxel-wise annotations of over 993,000 anatomical structures, covering pancreatic tumors, pancreas head, body, and tail, and 24 surrounding anatomical structures such as vascular/skeletal structures and abdominal/thoracic organs. Each scan includes metadata such as patient age, sex, diagnosis, contrast phase, in-plane spacing, slice thickness, etc. AI models trained on PanTS achieve significantly better performance in pancreatic tumor detection, localization, and segmentation compared to those trained on existing public datasets. Our analysis indicates that these gains are directly attributable to the 16x larger-scale tumor annotations and indirectly supported by the 24 additional surrounding anatomical structures. As the largest and most comprehensive resource of its kind, PanTS offers a new benchmark for developing and evaluating AI models in pancreatic CT analysis.
Text2CT: Towards 3D CT Volume Generation from Free-text Descriptions Using Diffusion Model
May 07, 2025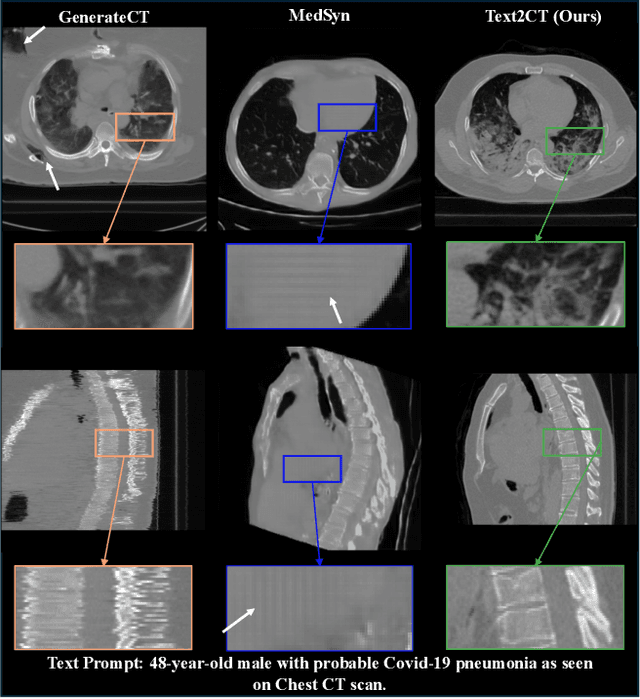
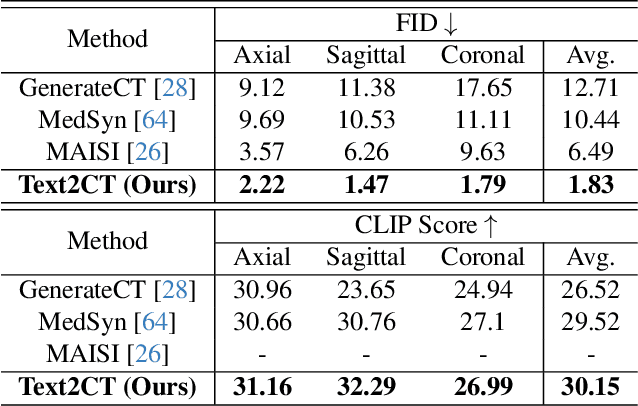
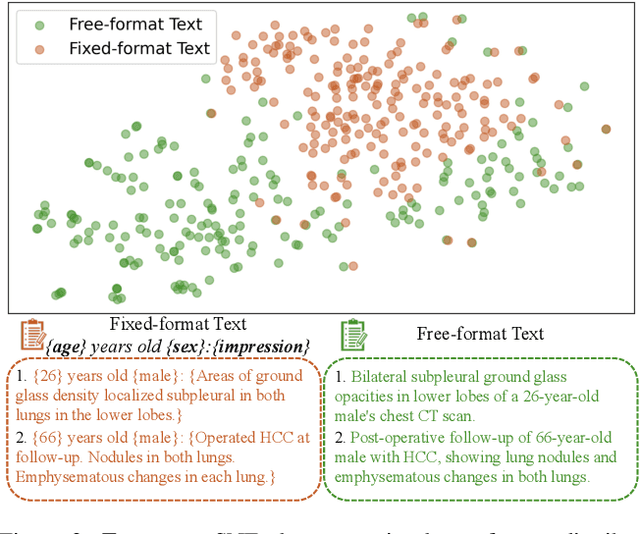
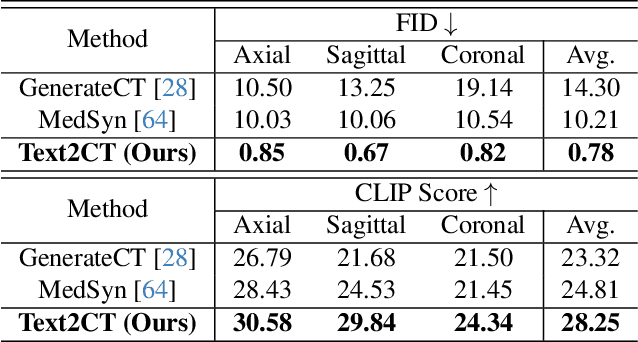
Abstract:Generating 3D CT volumes from descriptive free-text inputs presents a transformative opportunity in diagnostics and research. In this paper, we introduce Text2CT, a novel approach for synthesizing 3D CT volumes from textual descriptions using the diffusion model. Unlike previous methods that rely on fixed-format text input, Text2CT employs a novel prompt formulation that enables generation from diverse, free-text descriptions. The proposed framework encodes medical text into latent representations and decodes them into high-resolution 3D CT scans, effectively bridging the gap between semantic text inputs and detailed volumetric representations in a unified 3D framework. Our method demonstrates superior performance in preserving anatomical fidelity and capturing intricate structures as described in the input text. Extensive evaluations show that our approach achieves state-of-the-art results, offering promising potential applications in diagnostics, and data augmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge