Holger R. Roth
NVIDIA, USA
Standardized Methods and Recommendations for Green Federated Learning
Jan 30, 2026Abstract:Federated learning (FL) enables collaborative model training over privacy-sensitive, distributed data, but its environmental impact is difficult to compare across studies due to inconsistent measurement boundaries and heterogeneous reporting. We present a practical carbon-accounting methodology for FL CO2e tracking using NVIDIA NVFlare and CodeCarbon for explicit, phase-aware tasks (initialization, per-round training, evaluation, and idle/coordination). To capture non-compute effects, we additionally estimate communication emissions from transmitted model-update sizes under a network-configurable energy model. We validate the proposed approach on two representative workloads: CIFAR-10 image classification and retinal optic disk segmentation. In CIFAR-10, controlled client-efficiency scenarios show that system-level slowdowns and coordination effects can contribute meaningfully to carbon footprint under an otherwise fixed FL protocol, increasing total CO2e by 8.34x (medium) and 21.73x (low) relative to the high-efficiency baseline. In retinal segmentation, swapping GPU tiers (H100 vs.\ V100) yields a consistent 1.7x runtime gap (290 vs. 503 minutes) while producing non-uniform changes in total energy and CO2e across sites, underscoring the need for per-site and per-round reporting. Overall, our results support a standardized carbon accounting method that acts as a prerequisite for reproducible 'green' FL evaluation. Our code is available at https://github.com/Pediatric-Accelerated-Intelligence-Lab/carbon_footprint.
FeTTL: Federated Template and Task Learning for Multi-Institutional Medical Imaging
Jan 22, 2026Abstract:Federated learning enables collaborative model training across geographically distributed medical centers while preserving data privacy. However, domain shifts and heterogeneity in data often lead to a degradation in model performance. Medical imaging applications are particularly affected by variations in acquisition protocols, scanner types, and patient populations. To address these issues, we introduce Federated Template and Task Learning (FeTTL), a novel framework designed to harmonize multi-institutional medical imaging data in federated environments. FeTTL learns a global template together with a task model to align data distributions among clients. We evaluated FeTTL on two challenging and diverse multi-institutional medical imaging tasks: retinal fundus optical disc segmentation and histopathological metastasis classification. Experimental results show that FeTTL significantly outperforms the state-of-the-art federated learning baselines (p-values <0.002) for optical disc segmentation and classification of metastases from multi-institutional data. Our experiments further highlight the importance of jointly learning the template and the task. These findings suggest that FeTTL offers a principled and extensible solution for mitigating distribution shifts in federated learning, supporting robust model deployment in real-world, multi-institutional environments.
Federated Learning with Partially Labeled Data: A Conditional Distillation Approach
Dec 25, 2024



Abstract:In medical imaging, developing generalized segmentation models that can handle multiple organs and lesions is crucial. However, the scarcity of fully annotated datasets and strict privacy regulations present significant barriers to data sharing. Federated Learning (FL) allows decentralized model training, but existing FL methods often struggle with partial labeling, leading to model divergence and catastrophic forgetting. We propose ConDistFL, a novel FL framework incorporating conditional distillation to address these challenges. ConDistFL enables effective learning from partially labeled datasets, significantly improving segmentation accuracy across distributed and non-uniform datasets. In addition to its superior segmentation performance, ConDistFL maintains computational and communication efficiency, ensuring its scalability for real-world applications. Furthermore, ConDistFL demonstrates remarkable generalizability, significantly outperforming existing FL methods in out-of-federation tests, even adapting to unseen contrast phases (e.g., non-contrast CT images) in our experiments. Extensive evaluations on 3D CT and 2D chest X-ray datasets show that ConDistFL is an efficient, adaptable solution for collaborative medical image segmentation in privacy-constrained settings.
C-FedRAG: A Confidential Federated Retrieval-Augmented Generation System
Dec 17, 2024



Abstract:Organizations seeking to utilize Large Language Models (LLMs) for knowledge querying and analysis often encounter challenges in maintaining an LLM fine-tuned on targeted, up-to-date information that keeps answers relevant and grounded. Retrieval Augmented Generation (RAG) has quickly become a feasible solution for organizations looking to overcome the challenges of maintaining proprietary models and to help reduce LLM hallucinations in their query responses. However, RAG comes with its own issues regarding scaling data pipelines across tiered-access and disparate data sources. In many scenarios, it is necessary to query beyond a single data silo to provide richer and more relevant context for an LLM. Analyzing data sources within and across organizational trust boundaries is often limited by complex data-sharing policies that prohibit centralized data storage, therefore, inhibit the fast and effective setup and scaling of RAG solutions. In this paper, we introduce Confidential Computing (CC) techniques as a solution for secure Federated Retrieval Augmented Generation (FedRAG). Our proposed Confidential FedRAG system (C-FedRAG) enables secure connection and scaling of a RAG workflows across a decentralized network of data providers by ensuring context confidentiality. We also demonstrate how to implement a C-FedRAG system using the NVIDIA FLARE SDK and assess its performance using the MedRAG toolkit and MIRAGE benchmarking dataset.
Data Alchemy: Mitigating Cross-Site Model Variability Through Test Time Data Calibration
Jul 18, 2024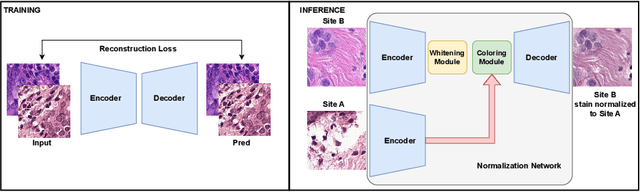
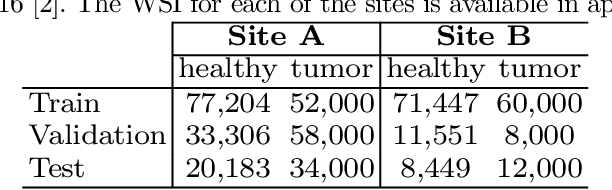
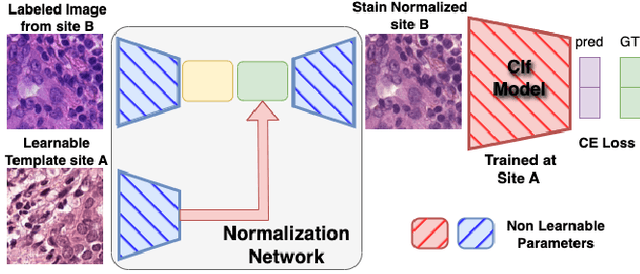
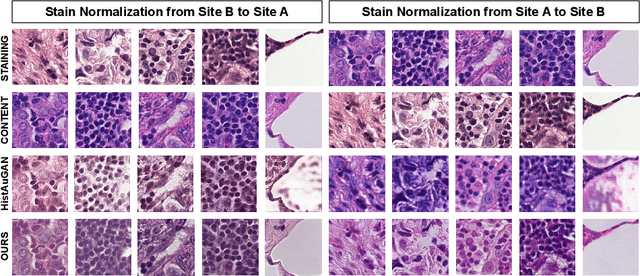
Abstract:Deploying deep learning-based imaging tools across various clinical sites poses significant challenges due to inherent domain shifts and regulatory hurdles associated with site-specific fine-tuning. For histopathology, stain normalization techniques can mitigate discrepancies, but they often fall short of eliminating inter-site variations. Therefore, we present Data Alchemy, an explainable stain normalization method combined with test time data calibration via a template learning framework to overcome barriers in cross-site analysis. Data Alchemy handles shifts inherent to multi-site data and minimizes them without needing to change the weights of the normalization or classifier networks. Our approach extends to unseen sites in various clinical settings where data domain discrepancies are unknown. Extensive experiments highlight the efficacy of our framework in tumor classification in hematoxylin and eosin-stained patches. Our explainable normalization method boosts classification tasks' area under the precision-recall curve(AUPR) by 0.165, 0.545 to 0.710. Additionally, Data Alchemy further reduces the multisite classification domain gap, by improving the 0.710 AUPR an additional 0.142, elevating classification performance further to 0.852, from 0.545. Our Data Alchemy framework can popularize precision medicine with minimal operational overhead by allowing for the seamless integration of pre-trained deep learning-based clinical tools across multiple sites.
D-Rax: Domain-specific Radiologic assistant leveraging multi-modal data and eXpert model predictions
Jul 02, 2024



Abstract:Large vision language models (VLMs) have progressed incredibly from research to applicability for general-purpose use cases. LLaVA-Med, a pioneering large language and vision assistant for biomedicine, can perform multi-modal biomedical image and data analysis to provide a natural language interface for radiologists. While it is highly generalizable and works with multi-modal data, it is currently limited by well-known challenges that exist in the large language model space. Hallucinations and imprecision in responses can lead to misdiagnosis which currently hinder the clinical adaptability of VLMs. To create precise, user-friendly models in healthcare, we propose D-Rax -- a domain-specific, conversational, radiologic assistance tool that can be used to gain insights about a particular radiologic image. In this study, we enhance the conversational analysis of chest X-ray (CXR) images to support radiological reporting, offering comprehensive insights from medical imaging and aiding in the formulation of accurate diagnosis. D-Rax is achieved by fine-tuning the LLaVA-Med architecture on our curated enhanced instruction-following data, comprising of images, instructions, as well as disease diagnosis and demographic predictions derived from MIMIC-CXR imaging data, CXR-related visual question answer (VQA) pairs, and predictive outcomes from multiple expert AI models. We observe statistically significant improvement in responses when evaluated for both open and close-ended conversations. Leveraging the power of state-of-the-art diagnostic models combined with VLMs, D-Rax empowers clinicians to interact with medical images using natural language, which could potentially streamline their decision-making process, enhance diagnostic accuracy, and conserve their time.
Fair Evaluation of Federated Learning Algorithms for Automated Breast Density Classification: The Results of the 2022 ACR-NCI-NVIDIA Federated Learning Challenge
May 22, 2024Abstract:The correct interpretation of breast density is important in the assessment of breast cancer risk. AI has been shown capable of accurately predicting breast density, however, due to the differences in imaging characteristics across mammography systems, models built using data from one system do not generalize well to other systems. Though federated learning (FL) has emerged as a way to improve the generalizability of AI without the need to share data, the best way to preserve features from all training data during FL is an active area of research. To explore FL methodology, the breast density classification FL challenge was hosted in partnership with the American College of Radiology, Harvard Medical School's Mass General Brigham, University of Colorado, NVIDIA, and the National Institutes of Health National Cancer Institute. Challenge participants were able to submit docker containers capable of implementing FL on three simulated medical facilities, each containing a unique large mammography dataset. The breast density FL challenge ran from June 15 to September 5, 2022, attracting seven finalists from around the world. The winning FL submission reached a linear kappa score of 0.653 on the challenge test data and 0.413 on an external testing dataset, scoring comparably to a model trained on the same data in a central location.
* 16 pages, 9 figures
Federated Learning Privacy: Attacks, Defenses, Applications, and Policy Landscape - A Survey
May 06, 2024Abstract:Deep learning has shown incredible potential across a vast array of tasks and accompanying this growth has been an insatiable appetite for data. However, a large amount of data needed for enabling deep learning is stored on personal devices and recent concerns on privacy have further highlighted challenges for accessing such data. As a result, federated learning (FL) has emerged as an important privacy-preserving technology enabling collaborative training of machine learning models without the need to send the raw, potentially sensitive, data to a central server. However, the fundamental premise that sending model updates to a server is privacy-preserving only holds if the updates cannot be "reverse engineered" to infer information about the private training data. It has been shown under a wide variety of settings that this premise for privacy does {\em not} hold. In this survey paper, we provide a comprehensive literature review of the different privacy attacks and defense methods in FL. We identify the current limitations of these attacks and highlight the settings in which FL client privacy can be broken. We dissect some of the successful industry applications of FL and draw lessons for future successful adoption. We survey the emerging landscape of privacy regulation for FL. We conclude with future directions for taking FL toward the cherished goal of generating accurate models while preserving the privacy of the data from its participants.
Empowering Federated Learning for Massive Models with NVIDIA FLARE
Feb 12, 2024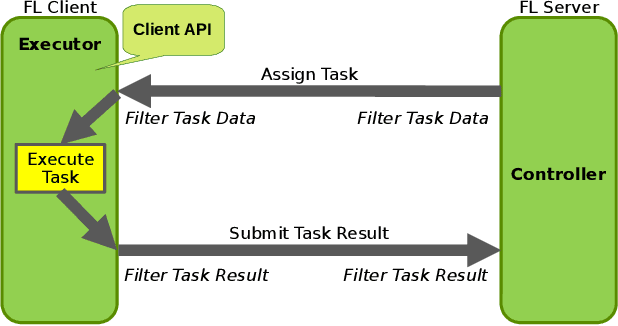

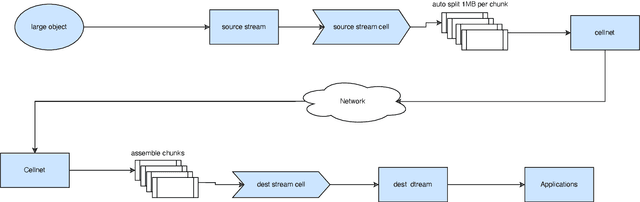
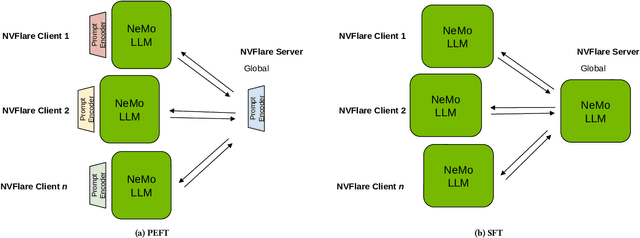
Abstract:In the ever-evolving landscape of artificial intelligence (AI) and large language models (LLMs), handling and leveraging data effectively has become a critical challenge. Most state-of-the-art machine learning algorithms are data-centric. However, as the lifeblood of model performance, necessary data cannot always be centralized due to various factors such as privacy, regulation, geopolitics, copyright issues, and the sheer effort required to move vast datasets. In this paper, we explore how federated learning enabled by NVIDIA FLARE can address these challenges with easy and scalable integration capabilities, enabling parameter-efficient and full supervised fine-tuning of LLMs for natural language processing and biopharmaceutical applications to enhance their accuracy and robustness.
FedBPT: Efficient Federated Black-box Prompt Tuning for Large Language Models
Oct 02, 2023



Abstract:Pre-trained language models (PLM) have revolutionized the NLP landscape, achieving stellar performances across diverse tasks. These models, while benefiting from vast training data, often require fine-tuning on specific data to cater to distinct downstream tasks. However, this data adaptation process has inherent security and privacy concerns, primarily when leveraging user-generated, device-residing data. Federated learning (FL) provides a solution, allowing collaborative model fine-tuning without centralized data collection. However, applying FL to finetune PLMs is hampered by challenges, including restricted model parameter access, high computational requirements, and communication overheads. This paper introduces Federated Black-box Prompt Tuning (FedBPT), a framework designed to address these challenges. FedBPT does not require the clients to access the model parameters. By focusing on training optimal prompts and utilizing gradient-free optimization methods, FedBPT reduces the number of exchanged variables, boosts communication efficiency, and minimizes computational and storage costs. Experiments highlight the framework's ability to drastically cut communication and memory costs while maintaining competitive performance. Ultimately, FedBPT presents a promising solution for efficient, privacy-preserving fine-tuning of PLM in the age of large language models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge