Pochuan Wang
Federated Learning with Partially Labeled Data: A Conditional Distillation Approach
Dec 25, 2024



Abstract:In medical imaging, developing generalized segmentation models that can handle multiple organs and lesions is crucial. However, the scarcity of fully annotated datasets and strict privacy regulations present significant barriers to data sharing. Federated Learning (FL) allows decentralized model training, but existing FL methods often struggle with partial labeling, leading to model divergence and catastrophic forgetting. We propose ConDistFL, a novel FL framework incorporating conditional distillation to address these challenges. ConDistFL enables effective learning from partially labeled datasets, significantly improving segmentation accuracy across distributed and non-uniform datasets. In addition to its superior segmentation performance, ConDistFL maintains computational and communication efficiency, ensuring its scalability for real-world applications. Furthermore, ConDistFL demonstrates remarkable generalizability, significantly outperforming existing FL methods in out-of-federation tests, even adapting to unseen contrast phases (e.g., non-contrast CT images) in our experiments. Extensive evaluations on 3D CT and 2D chest X-ray datasets show that ConDistFL is an efficient, adaptable solution for collaborative medical image segmentation in privacy-constrained settings.
ConDistFL: Conditional Distillation for Federated Learning from Partially Annotated Data
Aug 08, 2023



Abstract:Developing a generalized segmentation model capable of simultaneously delineating multiple organs and diseases is highly desirable. Federated learning (FL) is a key technology enabling the collaborative development of a model without exchanging training data. However, the limited access to fully annotated training data poses a major challenge to training generalizable models. We propose "ConDistFL", a framework to solve this problem by combining FL with knowledge distillation. Local models can extract the knowledge of unlabeled organs and tumors from partially annotated data from the global model with an adequately designed conditional probability representation. We validate our framework on four distinct partially annotated abdominal CT datasets from the MSD and KiTS19 challenges. The experimental results show that the proposed framework significantly outperforms FedAvg and FedOpt baselines. Moreover, the performance on an external test dataset demonstrates superior generalizability compared to models trained on each dataset separately. Our ablation study suggests that ConDistFL can perform well without frequent aggregation, reducing the communication cost of FL. Our implementation will be available at https://github.com/NVIDIA/NVFlare/tree/dev/research/condist-fl.
Multi-task Federated Learning for Heterogeneous Pancreas Segmentation
Aug 19, 2021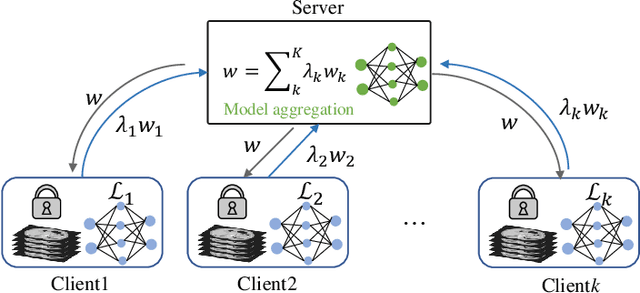
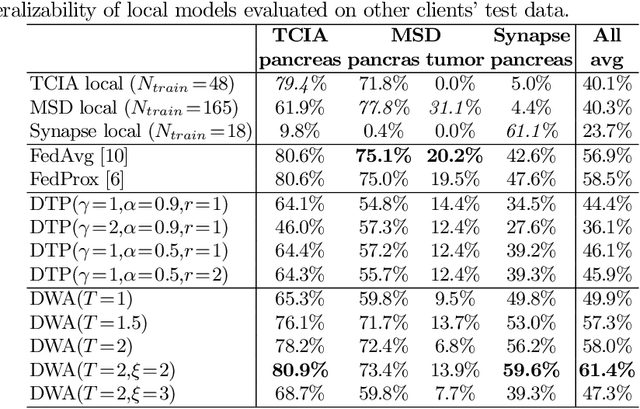
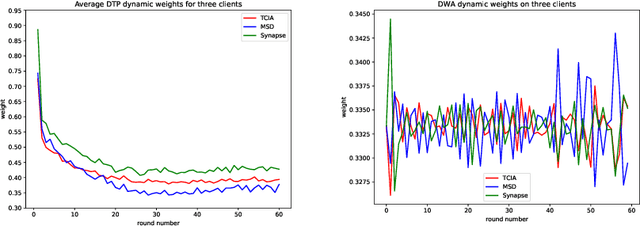
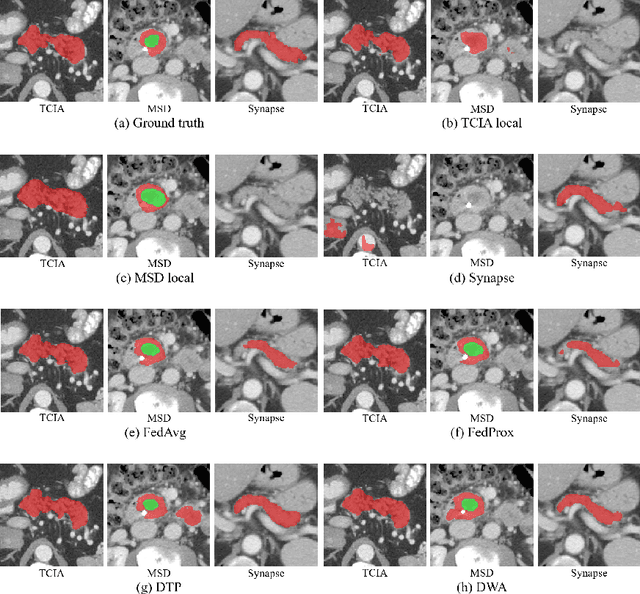
Abstract:Federated learning (FL) for medical image segmentation becomes more challenging in multi-task settings where clients might have different categories of labels represented in their data. For example, one client might have patient data with "healthy'' pancreases only while datasets from other clients may contain cases with pancreatic tumors. The vanilla federated averaging algorithm makes it possible to obtain more generalizable deep learning-based segmentation models representing the training data from multiple institutions without centralizing datasets. However, it might be sub-optimal for the aforementioned multi-task scenarios. In this paper, we investigate heterogeneous optimization methods that show improvements for the automated segmentation of pancreas and pancreatic tumors in abdominal CT images with FL settings.
Automated Pancreas Segmentation Using Multi-institutional Collaborative Deep Learning
Sep 28, 2020



Abstract:The performance of deep learning-based methods strongly relies on the number of datasets used for training. Many efforts have been made to increase the data in the medical image analysis field. However, unlike photography images, it is hard to generate centralized databases to collect medical images because of numerous technical, legal, and privacy issues. In this work, we study the use of federated learning between two institutions in a real-world setting to collaboratively train a model without sharing the raw data across national boundaries. We quantitatively compare the segmentation models obtained with federated learning and local training alone. Our experimental results show that federated learning models have higher generalizability than standalone training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge