Masahiro Oda
Federated Learning with Partially Labeled Data: A Conditional Distillation Approach
Dec 25, 2024



Abstract:In medical imaging, developing generalized segmentation models that can handle multiple organs and lesions is crucial. However, the scarcity of fully annotated datasets and strict privacy regulations present significant barriers to data sharing. Federated Learning (FL) allows decentralized model training, but existing FL methods often struggle with partial labeling, leading to model divergence and catastrophic forgetting. We propose ConDistFL, a novel FL framework incorporating conditional distillation to address these challenges. ConDistFL enables effective learning from partially labeled datasets, significantly improving segmentation accuracy across distributed and non-uniform datasets. In addition to its superior segmentation performance, ConDistFL maintains computational and communication efficiency, ensuring its scalability for real-world applications. Furthermore, ConDistFL demonstrates remarkable generalizability, significantly outperforming existing FL methods in out-of-federation tests, even adapting to unseen contrast phases (e.g., non-contrast CT images) in our experiments. Extensive evaluations on 3D CT and 2D chest X-ray datasets show that ConDistFL is an efficient, adaptable solution for collaborative medical image segmentation in privacy-constrained settings.
A Bayesian Approach to Weakly-supervised Laparoscopic Image Segmentation
Oct 11, 2024Abstract:In this paper, we study weakly-supervised laparoscopic image segmentation with sparse annotations. We introduce a novel Bayesian deep learning approach designed to enhance both the accuracy and interpretability of the model's segmentation, founded upon a comprehensive Bayesian framework, ensuring a robust and theoretically validated method. Our approach diverges from conventional methods that directly train using observed images and their corresponding weak annotations. Instead, we estimate the joint distribution of both images and labels given the acquired data. This facilitates the sampling of images and their high-quality pseudo-labels, enabling the training of a generalizable segmentation model. Each component of our model is expressed through probabilistic formulations, providing a coherent and interpretable structure. This probabilistic nature benefits accurate and practical learning from sparse annotations and equips our model with the ability to quantify uncertainty. Extensive evaluations with two public laparoscopic datasets demonstrated the efficacy of our method, which consistently outperformed existing methods. Furthermore, our method was adapted for scribble-supervised cardiac multi-structure segmentation, presenting competitive performance compared to previous methods. The code is available at https://github.com/MoriLabNU/Bayesian_WSS.
ConDistFL: Conditional Distillation for Federated Learning from Partially Annotated Data
Aug 08, 2023



Abstract:Developing a generalized segmentation model capable of simultaneously delineating multiple organs and diseases is highly desirable. Federated learning (FL) is a key technology enabling the collaborative development of a model without exchanging training data. However, the limited access to fully annotated training data poses a major challenge to training generalizable models. We propose "ConDistFL", a framework to solve this problem by combining FL with knowledge distillation. Local models can extract the knowledge of unlabeled organs and tumors from partially annotated data from the global model with an adequately designed conditional probability representation. We validate our framework on four distinct partially annotated abdominal CT datasets from the MSD and KiTS19 challenges. The experimental results show that the proposed framework significantly outperforms FedAvg and FedOpt baselines. Moreover, the performance on an external test dataset demonstrates superior generalizability compared to models trained on each dataset separately. Our ablation study suggests that ConDistFL can perform well without frequent aggregation, reducing the communication cost of FL. Our implementation will be available at https://github.com/NVIDIA/NVFlare/tree/dev/research/condist-fl.
Identifying Suspicious Regions of Covid-19 by Abnormality-Sensitive Activation Mapping
Mar 27, 2023Abstract:This paper presents a fully-automated method for the identification of suspicious regions of a coronavirus disease (COVID-19) on chest CT volumes. One major role of chest CT scanning in COVID-19 diagnoses is identification of an inflammation particular to the disease. This task is generally performed by radiologists through an interpretation of the CT volumes, however, because of the heavy workload, an automatic analysis method using a computer is desired. Most computer-aided diagnosis studies have addressed only a portion of the elements necessary for the identification. In this work, we realize the identification method through a classification task by using a 2.5-dimensional CNN with three-dimensional attention mechanisms. We visualize the suspicious regions by applying a backpropagation based on positive gradients to attention-weighted features. We perform experiments on an in-house dataset and two public datasets to reveal the generalization ability of the proposed method. The proposed architecture achieved AUCs of over 0.900 for all the datasets, and mean sensitivity $0.853 \pm 0.036$ and specificity $0.870 \pm 0.040$. The method can also identify notable lesions pointed out in the radiology report as suspicious regions.
KST-Mixer: Kinematic Spatio-Temporal Data Mixer For Colon Shape Estimation
Feb 02, 2023Abstract:We propose a spatio-temporal mixing kinematic data estimation method to estimate the shape of the colon with deformations caused by colonoscope insertion. Endoscope tracking or a navigation system that navigates physicians to target positions is needed to reduce such complications as organ perforations. Although many previous methods focused to track bronchoscopes and surgical endoscopes, few number of colonoscope tracking methods were proposed. This is because the colon largely deforms during colonoscope insertion. The deformation causes significant tracking errors. Colon deformation should be taken into account in the tracking process. We propose a colon shape estimation method using a Kinematic Spatio-Temporal data Mixer (KST-Mixer) that can be used during colonoscope insertions to the colon. Kinematic data of a colonoscope and the colon, including positions and directions of their centerlines, are obtained using electromagnetic and depth sensors. The proposed method separates the data into sub-groups along the spatial and temporal axes. The KST-Mixer extracts kinematic features and mix them along the spatial and temporal axes multiple times. We evaluated colon shape estimation accuracies in phantom studies. The proposed method achieved 11.92 mm mean Euclidean distance error, the smallest of the previous methods. Statistical analysis indicated that the proposed method significantly reduced the error compared to the previous methods.
* Accepted paper as an oral presentation at Joint MICCAI workshop 2022, AE-CAI/CARE/OR2.0. Received the Outstanding Paper Award
Semi-automated Virtual Unfolded View Generation Method of Stomach from CT Volumes
Jan 14, 2022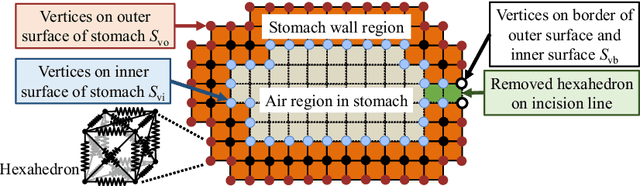
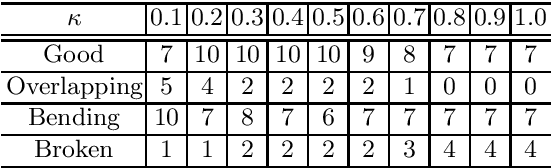
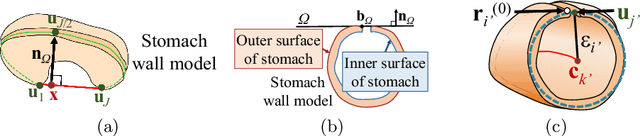

Abstract:CT image-based diagnosis of the stomach is developed as a new way of diagnostic method. A virtual unfolded (VU) view is suitable for displaying its wall. In this paper, we propose a semi-automated method for generating VU views of the stomach. Our method requires minimum manual operations. The determination of the unfolding forces and the termination of the unfolding process are automated. The unfolded shape of the stomach is estimated based on its radius. The unfolding forces are determined so that the stomach wall is deformed to the expected shape. The iterative deformation process is terminated if the difference of the shapes between the deformed shape and expected shape is small. Our experiments using 67 CT volumes showed that our proposed method can generate good VU views for 76.1% cases.
* Accepted paper as a poster presentation at MICCAI 2013 (International Conference on Medical Image Computing and Computer-Assisted Intervention), Nagoya, Japan
Realistic Endoscopic Image Generation Method Using Virtual-to-real Image-domain Translation
Jan 13, 2022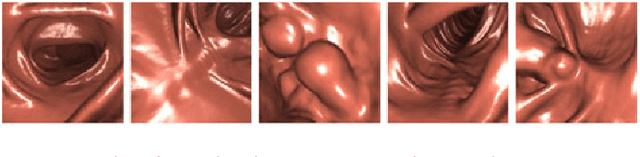

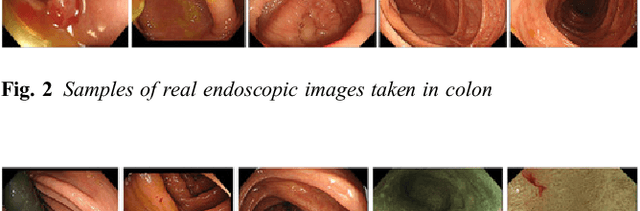
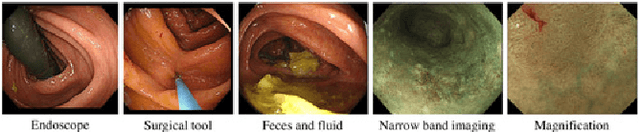
Abstract:This paper proposes a realistic image generation method for visualization in endoscopic simulation systems. Endoscopic diagnosis and treatment are performed in many hospitals. To reduce complications related to endoscope insertions, endoscopic simulation systems are used for training or rehearsal of endoscope insertions. However, current simulation systems generate non-realistic virtual endoscopic images. To improve the value of the simulation systems, improvement of reality of their generated images is necessary. We propose a realistic image generation method for endoscopic simulation systems. Virtual endoscopic images are generated by using a volume rendering method from a CT volume of a patient. We improve the reality of the virtual endoscopic images using a virtual-to-real image-domain translation technique. The image-domain translator is implemented as a fully convolutional network (FCN). We train the FCN by minimizing a cycle consistency loss function. The FCN is trained using unpaired virtual and real endoscopic images. To obtain high quality image-domain translation results, we perform an image cleansing to the real endoscopic image set. We tested to use the shallow U-Net, U-Net, deep U-Net, and U-Net having residual units as the image-domain translator. The deep U-Net and U-Net having residual units generated quite realistic images.
* Accepted paper as an oral presentation at the Joint MICCAI workshop MIAR | AE-CAI | CARE 2019
Depth Estimation from Single-shot Monocular Endoscope Image Using Image Domain Adaptation And Edge-Aware Depth Estimation
Jan 12, 2022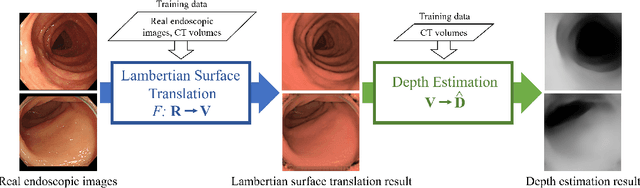

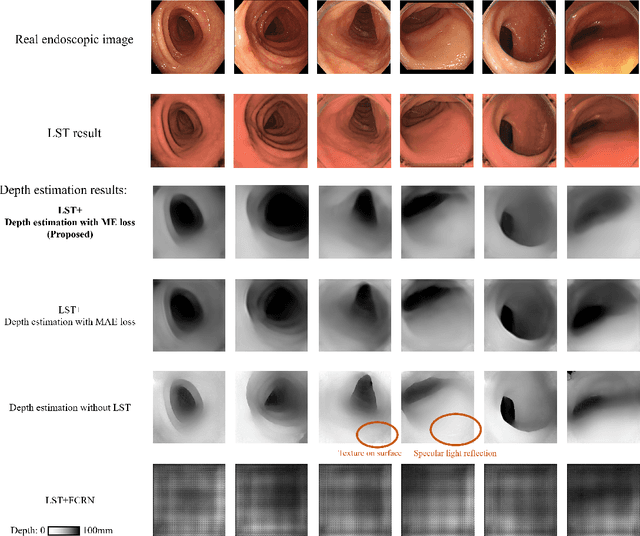

Abstract:We propose a depth estimation method from a single-shot monocular endoscopic image using Lambertian surface translation by domain adaptation and depth estimation using multi-scale edge loss. We employ a two-step estimation process including Lambertian surface translation from unpaired data and depth estimation. The texture and specular reflection on the surface of an organ reduce the accuracy of depth estimations. We apply Lambertian surface translation to an endoscopic image to remove these texture and reflections. Then, we estimate the depth by using a fully convolutional network (FCN). During the training of the FCN, improvement of the object edge similarity between an estimated image and a ground truth depth image is important for getting better results. We introduced a muti-scale edge loss function to improve the accuracy of depth estimation. We quantitatively evaluated the proposed method using real colonoscopic images. The estimated depth values were proportional to the real depth values. Furthermore, we applied the estimated depth images to automated anatomical location identification of colonoscopic images using a convolutional neural network. The identification accuracy of the network improved from 69.2% to 74.1% by using the estimated depth images.
* Accepted paper as an oral presentation at Joint MICCAI workshop 2021, AE-CAI/CARE/OR2.0
COVID-19 Infection Segmentation from Chest CT Images Based on Scale Uncertainty
Jan 09, 2022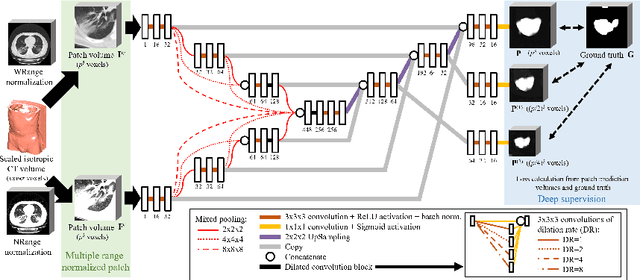
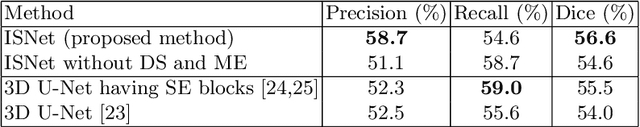
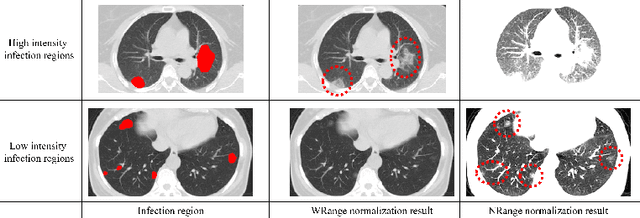

Abstract:This paper proposes a segmentation method of infection regions in the lung from CT volumes of COVID-19 patients. COVID-19 spread worldwide, causing many infected patients and deaths. CT image-based diagnosis of COVID-19 can provide quick and accurate diagnosis results. An automated segmentation method of infection regions in the lung provides a quantitative criterion for diagnosis. Previous methods employ whole 2D image or 3D volume-based processes. Infection regions have a considerable variation in their sizes. Such processes easily miss small infection regions. Patch-based process is effective for segmenting small targets. However, selecting the appropriate patch size is difficult in infection region segmentation. We utilize the scale uncertainty among various receptive field sizes of a segmentation FCN to obtain infection regions. The receptive field sizes can be defined as the patch size and the resolution of volumes where patches are clipped from. This paper proposes an infection segmentation network (ISNet) that performs patch-based segmentation and a scale uncertainty-aware prediction aggregation method that refines the segmentation result. We design ISNet to segment infection regions that have various intensity values. ISNet has multiple encoding paths to process patch volumes normalized by multiple intensity ranges. We collect prediction results generated by ISNets having various receptive field sizes. Scale uncertainty among the prediction results is extracted by the prediction aggregation method. We use an aggregation FCN to generate a refined segmentation result considering scale uncertainty among the predictions. In our experiments using 199 chest CT volumes of COVID-19 cases, the prediction aggregation method improved the dice similarity score from 47.6% to 62.1%.
* Accepted paper as a oral presentation at CILP2021, 10th MICCAI CLIP Workshop
Lung infection and normal region segmentation from CT volumes of COVID-19 cases
Jan 09, 2022Abstract:This paper proposes an automated segmentation method of infection and normal regions in the lung from CT volumes of COVID-19 patients. From December 2019, novel coronavirus disease 2019 (COVID-19) spreads over the world and giving significant impacts to our economic activities and daily lives. To diagnose the large number of infected patients, diagnosis assistance by computers is needed. Chest CT is effective for diagnosis of viral pneumonia including COVID-19. A quantitative analysis method of condition of the lung from CT volumes by computers is required for diagnosis assistance of COVID-19. This paper proposes an automated segmentation method of infection and normal regions in the lung from CT volumes using a COVID-19 segmentation fully convolutional network (FCN). In diagnosis of lung diseases including COVID-19, analysis of conditions of normal and infection regions in the lung is important. Our method recognizes and segments lung normal and infection regions in CT volumes. To segment infection regions that have various shapes and sizes, we introduced dense pooling connections and dilated convolutions in our FCN. We applied the proposed method to CT volumes of COVID-19 cases. From mild to severe cases of COVID-19, the proposed method correctly segmented normal and infection regions in the lung. Dice scores of normal and infection regions were 0.911 and 0.753, respectively.
* Accepted paper as a poster presentation at SPIE Medical Imaging 2021
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge