Kiyohito Tanaka
Domain Adaptation for Ulcerative Colitis Severity Estimation Using Patient-Level Diagnoses
Sep 18, 2025



Abstract:The development of methods to estimate the severity of Ulcerative Colitis (UC) is of significant importance. However, these methods often suffer from domain shifts caused by differences in imaging devices and clinical settings across hospitals. Although several domain adaptation methods have been proposed to address domain shift, they still struggle with the lack of supervision in the target domain or the high cost of annotation. To overcome these challenges, we propose a novel Weakly Supervised Domain Adaptation method that leverages patient-level diagnostic results, which are routinely recorded in UC diagnosis, as weak supervision in the target domain. The proposed method aligns class-wise distributions across domains using Shared Aggregation Tokens and a Max-Severity Triplet Loss, which leverages the characteristic that patient-level diagnoses are determined by the most severe region within each patient. Experimental results demonstrate that our method outperforms comparative DA approaches, improving UC severity estimation in a domain-shifted setting.
Ordinal Multiple-instance Learning for Ulcerative Colitis Severity Estimation with Selective Aggregated Transformer
Nov 22, 2024



Abstract:Patient-level diagnosis of severity in ulcerative colitis (UC) is common in real clinical settings, where the most severe score in a patient is recorded. However, previous UC classification methods (i.e., image-level estimation) mainly assumed the input was a single image. Thus, these methods can not utilize severity labels recorded in real clinical settings. In this paper, we propose a patient-level severity estimation method by a transformer with selective aggregator tokens, where a severity label is estimated from multiple images taken from a patient, similar to a clinical setting. Our method can effectively aggregate features of severe parts from a set of images captured in each patient, and it facilitates improving the discriminative ability between adjacent severity classes. Experiments demonstrate the effectiveness of the proposed method on two datasets compared with the state-of-the-art MIL methods. Moreover, we evaluated our method in real clinical settings and confirmed that our method outperformed the previous image-level methods. The code is publicly available at https://github.com/Shiku-Kaito/Ordinal-Multiple-instance-Learning-for-Ulcerative-Colitis-Severity-Estimation.
Self-Relaxed Joint Training: Sample Selection for Severity Estimation with Ordinal Noisy Labels
Oct 29, 2024



Abstract:Severity level estimation is a crucial task in medical image diagnosis. However, accurately assigning severity class labels to individual images is very costly and challenging. Consequently, the attached labels tend to be noisy. In this paper, we propose a new framework for training with ``ordinal'' noisy labels. Since severity levels have an ordinal relationship, we can leverage this to train a classifier while mitigating the negative effects of noisy labels. Our framework uses two techniques: clean sample selection and dual-network architecture. A technical highlight of our approach is the use of soft labels derived from noisy hard labels. By appropriately using the soft and hard labels in the two techniques, we achieve more accurate sample selection and robust network training. The proposed method outperforms various state-of-the-art methods in experiments using two endoscopic ulcerative colitis (UC) datasets and a retinal Diabetic Retinopathy (DR) dataset. Our codes are available at https://github.com/shumpei-takezaki/Self-Relaxed-Joint-Training.
Deep Bayesian Active Learning-to-Rank with Relative Annotation for Estimation of Ulcerative Colitis Severity
Sep 10, 2024



Abstract:Automatic image-based severity estimation is an important task in computer-aided diagnosis. Severity estimation by deep learning requires a large amount of training data to achieve a high performance. In general, severity estimation uses training data annotated with discrete (i.e., quantized) severity labels. Annotating discrete labels is often difficult in images with ambiguous severity, and the annotation cost is high. In contrast, relative annotation, in which the severity between a pair of images is compared, can avoid quantizing severity and thus makes it easier. We can estimate relative disease severity using a learning-to-rank framework with relative annotations, but relative annotation has the problem of the enormous number of pairs that can be annotated. Therefore, the selection of appropriate pairs is essential for relative annotation. In this paper, we propose a deep Bayesian active learning-to-rank that automatically selects appropriate pairs for relative annotation. Our method preferentially annotates unlabeled pairs with high learning efficiency from the model uncertainty of the samples. We prove the theoretical basis for adapting Bayesian neural networks to pairwise learning-to-rank and demonstrate the efficiency of our method through experiments on endoscopic images of ulcerative colitis on both private and public datasets. We also show that our method achieves a high performance under conditions of significant class imbalance because it automatically selects samples from the minority classes.
* 14 pages, 8 figures, accepted in Medical Image Analysis 2024
Disease Severity Regression with Continuous Data Augmentation
Feb 24, 2023



Abstract:Disease severity regression by a convolutional neural network (CNN) for medical images requires a sufficient number of image samples labeled with severity levels. Conditional generative adversarial network (cGAN)-based data augmentation (DA) is a possible solution, but it encounters two issues. The first issue is that existing cGANs cannot deal with real-valued severity levels as their conditions, and the second is that the severity of the generated images is not fully reliable. We propose continuous DA as a solution to the two issues. Our method uses continuous severity GAN to generate images at real-valued severity levels and dataset-disjoint multi-objective optimization to deal with the second issue. Our method was evaluated for estimating ulcerative colitis (UC) severity of endoscopic images and achieved higher classification performance than conventional DA methods.
Deep Bayesian Active-Learning-to-Rank for Endoscopic Image Data
Aug 05, 2022Abstract:Automatic image-based disease severity estimation generally uses discrete (i.e., quantized) severity labels. Annotating discrete labels is often difficult due to the images with ambiguous severity. An easier alternative is to use relative annotation, which compares the severity level between image pairs. By using a learning-to-rank framework with relative annotation, we can train a neural network that estimates rank scores that are relative to severity levels. However, the relative annotation for all possible pairs is prohibitive, and therefore, appropriate sample pair selection is mandatory. This paper proposes a deep Bayesian active-learning-to-rank, which trains a Bayesian convolutional neural network while automatically selecting appropriate pairs for relative annotation. We confirmed the efficiency of the proposed method through experiments on endoscopic images of ulcerative colitis. In addition, we confirmed that our method is useful even with the severe class imbalance because of its ability to select samples from minor classes automatically.
Realistic Endoscopic Image Generation Method Using Virtual-to-real Image-domain Translation
Jan 13, 2022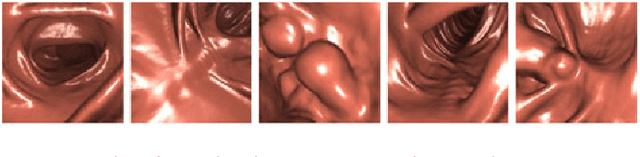

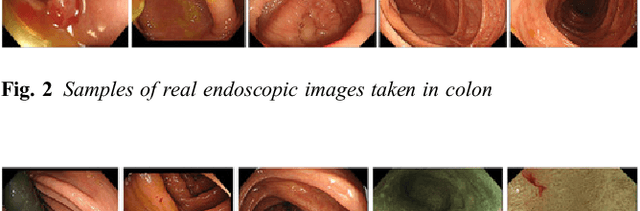
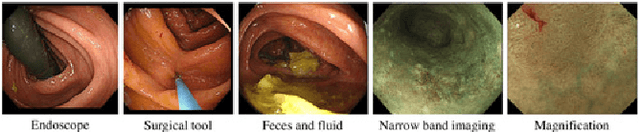
Abstract:This paper proposes a realistic image generation method for visualization in endoscopic simulation systems. Endoscopic diagnosis and treatment are performed in many hospitals. To reduce complications related to endoscope insertions, endoscopic simulation systems are used for training or rehearsal of endoscope insertions. However, current simulation systems generate non-realistic virtual endoscopic images. To improve the value of the simulation systems, improvement of reality of their generated images is necessary. We propose a realistic image generation method for endoscopic simulation systems. Virtual endoscopic images are generated by using a volume rendering method from a CT volume of a patient. We improve the reality of the virtual endoscopic images using a virtual-to-real image-domain translation technique. The image-domain translator is implemented as a fully convolutional network (FCN). We train the FCN by minimizing a cycle consistency loss function. The FCN is trained using unpaired virtual and real endoscopic images. To obtain high quality image-domain translation results, we perform an image cleansing to the real endoscopic image set. We tested to use the shallow U-Net, U-Net, deep U-Net, and U-Net having residual units as the image-domain translator. The deep U-Net and U-Net having residual units generated quite realistic images.
* Accepted paper as an oral presentation at the Joint MICCAI workshop MIAR | AE-CAI | CARE 2019
Depth Estimation from Single-shot Monocular Endoscope Image Using Image Domain Adaptation And Edge-Aware Depth Estimation
Jan 12, 2022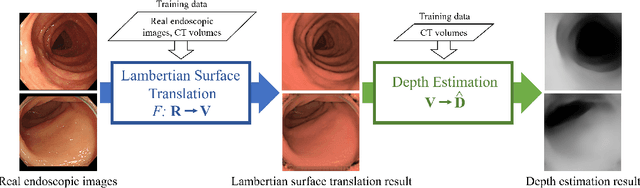

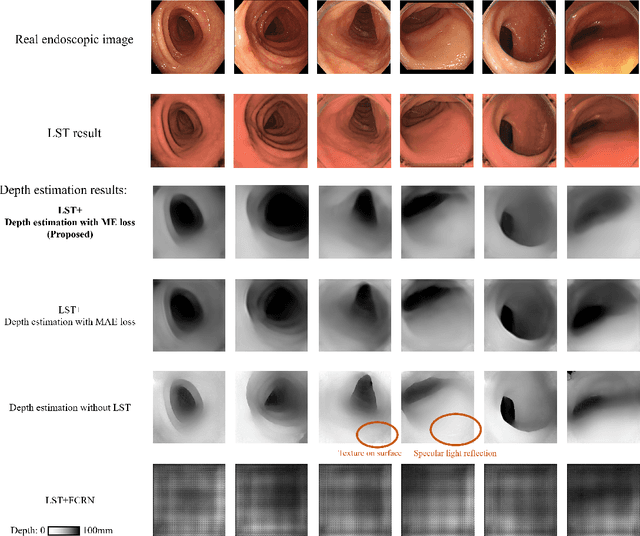

Abstract:We propose a depth estimation method from a single-shot monocular endoscopic image using Lambertian surface translation by domain adaptation and depth estimation using multi-scale edge loss. We employ a two-step estimation process including Lambertian surface translation from unpaired data and depth estimation. The texture and specular reflection on the surface of an organ reduce the accuracy of depth estimations. We apply Lambertian surface translation to an endoscopic image to remove these texture and reflections. Then, we estimate the depth by using a fully convolutional network (FCN). During the training of the FCN, improvement of the object edge similarity between an estimated image and a ground truth depth image is important for getting better results. We introduced a muti-scale edge loss function to improve the accuracy of depth estimation. We quantitatively evaluated the proposed method using real colonoscopic images. The estimated depth values were proportional to the real depth values. Furthermore, we applied the estimated depth images to automated anatomical location identification of colonoscopic images using a convolutional neural network. The identification accuracy of the network improved from 69.2% to 74.1% by using the estimated depth images.
* Accepted paper as an oral presentation at Joint MICCAI workshop 2021, AE-CAI/CARE/OR2.0
Order-Guided Disentangled Representation Learning for Ulcerative Colitis Classification with Limited Labels
Nov 06, 2021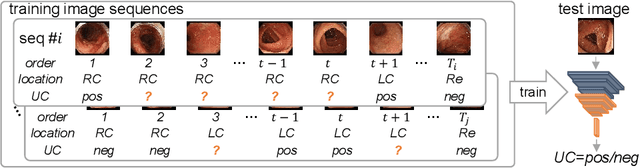
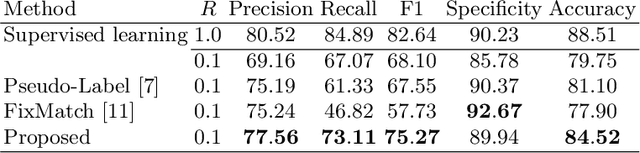

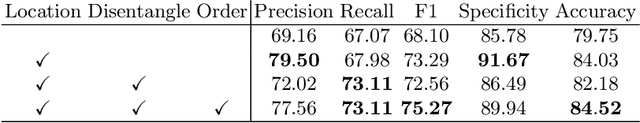
Abstract:Ulcerative colitis (UC) classification, which is an important task for endoscopic diagnosis, involves two main difficulties. First, endoscopic images with the annotation about UC (positive or negative) are usually limited. Second, they show a large variability in their appearance due to the location in the colon. Especially, the second difficulty prevents us from using existing semi-supervised learning techniques, which are the common remedy for the first difficulty. In this paper, we propose a practical semi-supervised learning method for UC classification by newly exploiting two additional features, the location in a colon (e.g., left colon) and image capturing order, both of which are often attached to individual images in endoscopic image sequences. The proposed method can extract the essential information of UC classification efficiently by a disentanglement process with those features. Experimental results demonstrate that the proposed method outperforms several existing semi-supervised learning methods in the classification task, even with a small number of annotated images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge