Hayato Itoh
Identifying Suspicious Regions of Covid-19 by Abnormality-Sensitive Activation Mapping
Mar 27, 2023Abstract:This paper presents a fully-automated method for the identification of suspicious regions of a coronavirus disease (COVID-19) on chest CT volumes. One major role of chest CT scanning in COVID-19 diagnoses is identification of an inflammation particular to the disease. This task is generally performed by radiologists through an interpretation of the CT volumes, however, because of the heavy workload, an automatic analysis method using a computer is desired. Most computer-aided diagnosis studies have addressed only a portion of the elements necessary for the identification. In this work, we realize the identification method through a classification task by using a 2.5-dimensional CNN with three-dimensional attention mechanisms. We visualize the suspicious regions by applying a backpropagation based on positive gradients to attention-weighted features. We perform experiments on an in-house dataset and two public datasets to reveal the generalization ability of the proposed method. The proposed architecture achieved AUCs of over 0.900 for all the datasets, and mean sensitivity $0.853 \pm 0.036$ and specificity $0.870 \pm 0.040$. The method can also identify notable lesions pointed out in the radiology report as suspicious regions.
Depth Estimation from Single-shot Monocular Endoscope Image Using Image Domain Adaptation And Edge-Aware Depth Estimation
Jan 12, 2022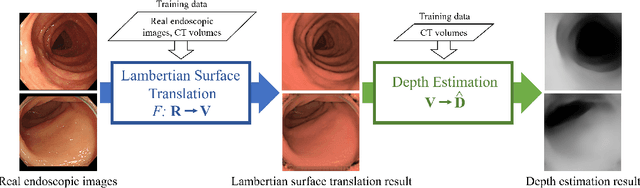

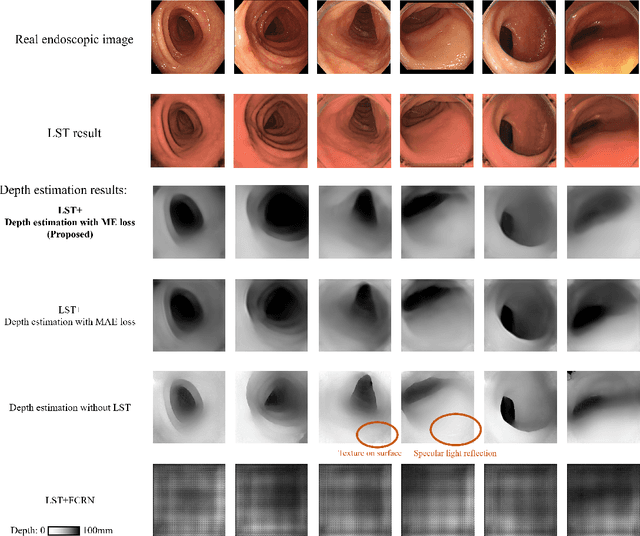

Abstract:We propose a depth estimation method from a single-shot monocular endoscopic image using Lambertian surface translation by domain adaptation and depth estimation using multi-scale edge loss. We employ a two-step estimation process including Lambertian surface translation from unpaired data and depth estimation. The texture and specular reflection on the surface of an organ reduce the accuracy of depth estimations. We apply Lambertian surface translation to an endoscopic image to remove these texture and reflections. Then, we estimate the depth by using a fully convolutional network (FCN). During the training of the FCN, improvement of the object edge similarity between an estimated image and a ground truth depth image is important for getting better results. We introduced a muti-scale edge loss function to improve the accuracy of depth estimation. We quantitatively evaluated the proposed method using real colonoscopic images. The estimated depth values were proportional to the real depth values. Furthermore, we applied the estimated depth images to automated anatomical location identification of colonoscopic images using a convolutional neural network. The identification accuracy of the network improved from 69.2% to 74.1% by using the estimated depth images.
* Accepted paper as an oral presentation at Joint MICCAI workshop 2021, AE-CAI/CARE/OR2.0
Visualizing intestines for diagnostic assistance of ileus based on intestinal region segmentation from 3D CT images
Mar 03, 2020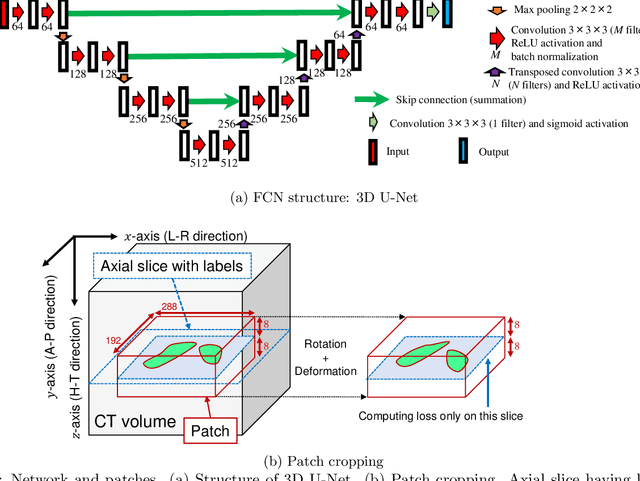
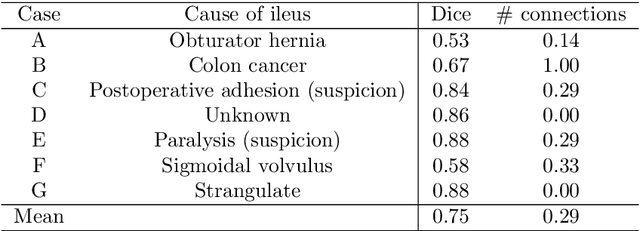
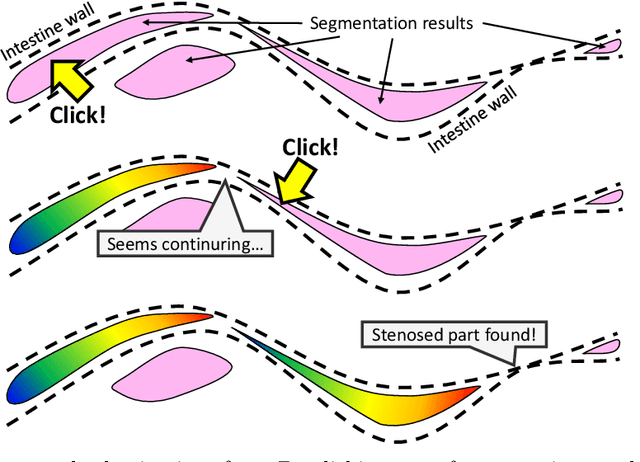
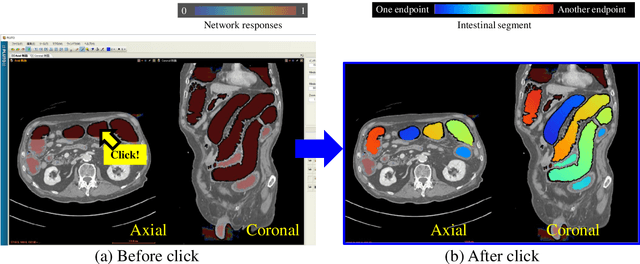
Abstract:This paper presents a visualization method of intestine (the small and large intestines) regions and their stenosed parts caused by ileus from CT volumes. Since it is difficult for non-expert clinicians to find stenosed parts, the intestine and its stenosed parts should be visualized intuitively. Furthermore, the intestine regions of ileus cases are quite hard to be segmented. The proposed method segments intestine regions by 3D FCN (3D U-Net). Intestine regions are quite difficult to be segmented in ileus cases since the inside the intestine is filled with fluids. These fluids have similar intensities with intestinal wall on 3D CT volumes. We segment the intestine regions by using 3D U-Net trained by a weak annotation approach. Weak-annotation makes possible to train the 3D U-Net with small manually-traced label images of the intestine. This avoids us to prepare many annotation labels of the intestine that has long and winding shape. Each intestine segment is volume-rendered and colored based on the distance from its endpoint in volume rendering. Stenosed parts (disjoint points of an intestine segment) can be easily identified on such visualization. In the experiments, we showed that stenosed parts were intuitively visualized as endpoints of segmented regions, which are colored by red or blue.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge