Hirohisa Oda
Micro CT Image-Assisted Cross Modality Super-Resolution of Clinical CT Images Utilizing Synthesized Training Dataset
Oct 20, 2020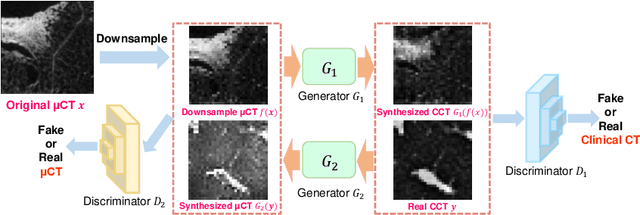
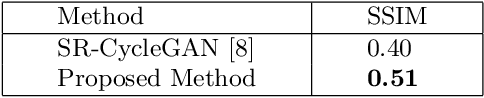
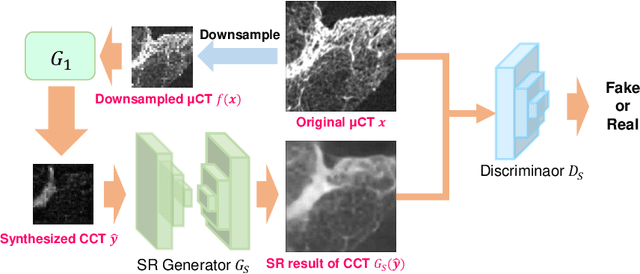

Abstract:This paper proposes a novel, unsupervised super-resolution (SR) approach for performing the SR of a clinical CT into the resolution level of a micro CT ($\mu$CT). The precise non-invasive diagnosis of lung cancer typically utilizes clinical CT data. Due to the resolution limitations of clinical CT (about $0.5 \times 0.5 \times 0.5$ mm$^3$), it is difficult to obtain enough pathological information such as the invasion area at alveoli level. On the other hand, $\mu$CT scanning allows the acquisition of volumes of lung specimens with much higher resolution ($50 \times 50 \times 50 \mu {\rm m}^3$ or higher). Thus, super-resolution of clinical CT volume may be helpful for diagnosis of lung cancer. Typical SR methods require aligned pairs of low-resolution (LR) and high-resolution (HR) images for training. Unfortunately, obtaining paired clinical CT and $\mu$CT volumes of human lung tissues is infeasible. Unsupervised SR methods are required that do not need paired LR and HR images. In this paper, we create corresponding clinical CT-$\mu$CT pairs by simulating clinical CT images from $\mu$CT images by modified CycleGAN. After this, we use simulated clinical CT-$\mu$CT image pairs to train an SR network based on SRGAN. Finally, we use the trained SR network to perform SR of the clinical CT images. We compare our proposed method with another unsupervised SR method for clinical CT images named SR-CycleGAN. Experimental results demonstrate that the proposed method can successfully perform SR of clinical CT images of lung cancer patients with $\mu$CT level resolution, and quantitatively and qualitatively outperformed conventional method (SR-CycleGAN), improving the SSIM (structure similarity) form 0.40 to 0.51.
Visualizing intestines for diagnostic assistance of ileus based on intestinal region segmentation from 3D CT images
Mar 03, 2020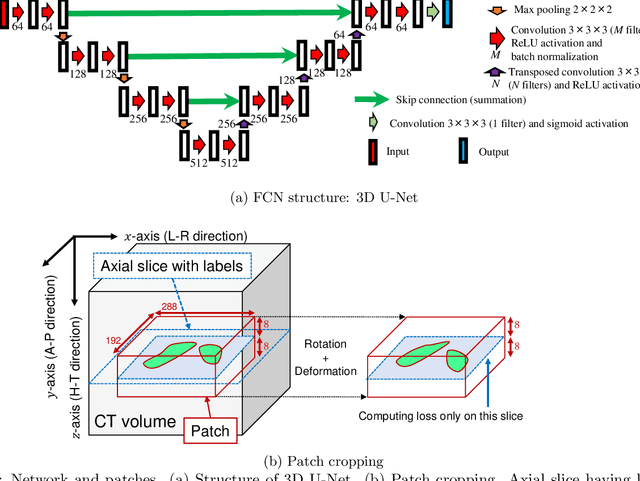
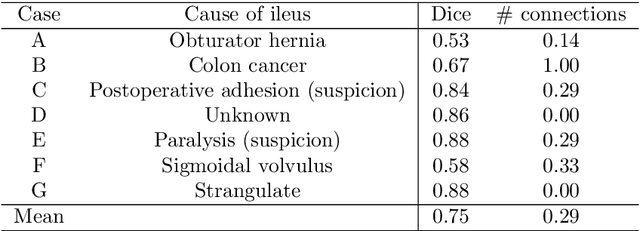
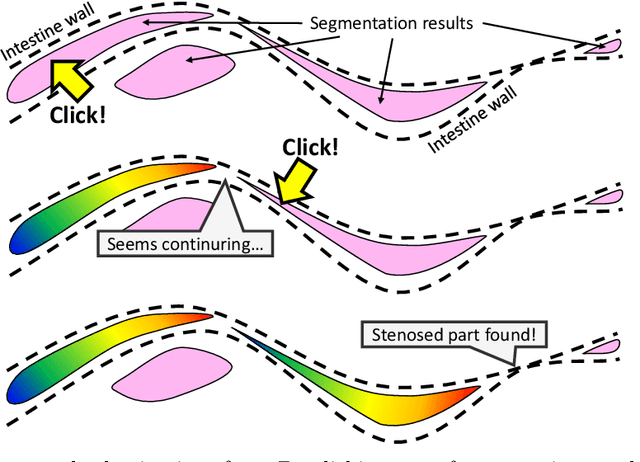
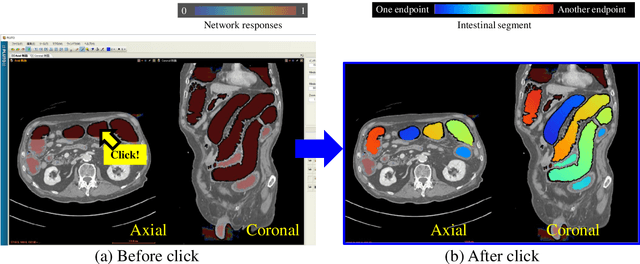
Abstract:This paper presents a visualization method of intestine (the small and large intestines) regions and their stenosed parts caused by ileus from CT volumes. Since it is difficult for non-expert clinicians to find stenosed parts, the intestine and its stenosed parts should be visualized intuitively. Furthermore, the intestine regions of ileus cases are quite hard to be segmented. The proposed method segments intestine regions by 3D FCN (3D U-Net). Intestine regions are quite difficult to be segmented in ileus cases since the inside the intestine is filled with fluids. These fluids have similar intensities with intestinal wall on 3D CT volumes. We segment the intestine regions by using 3D U-Net trained by a weak annotation approach. Weak-annotation makes possible to train the 3D U-Net with small manually-traced label images of the intestine. This avoids us to prepare many annotation labels of the intestine that has long and winding shape. Each intestine segment is volume-rendered and colored based on the distance from its endpoint in volume rendering. Stenosed parts (disjoint points of an intestine segment) can be easily identified on such visualization. In the experiments, we showed that stenosed parts were intuitively visualized as endpoints of segmented regions, which are colored by red or blue.
Multi-modality super-resolution loss for GAN-based super-resolution of clinical CT images using micro CT image database
Dec 30, 2019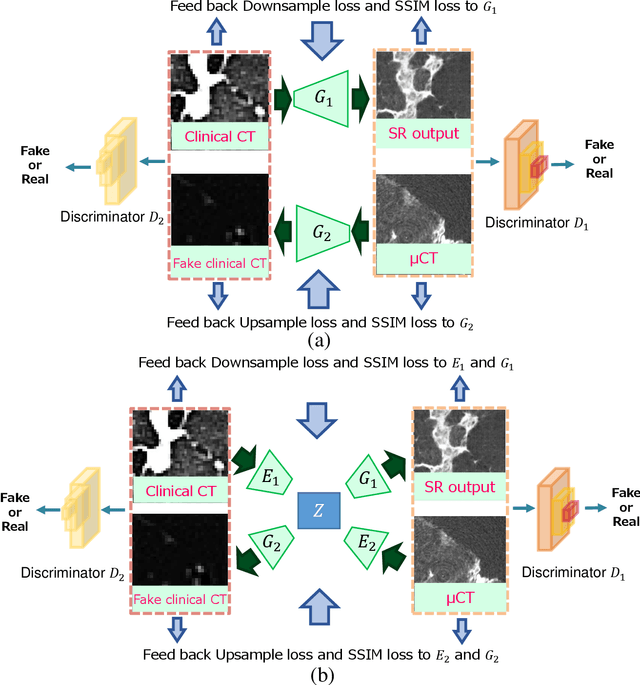
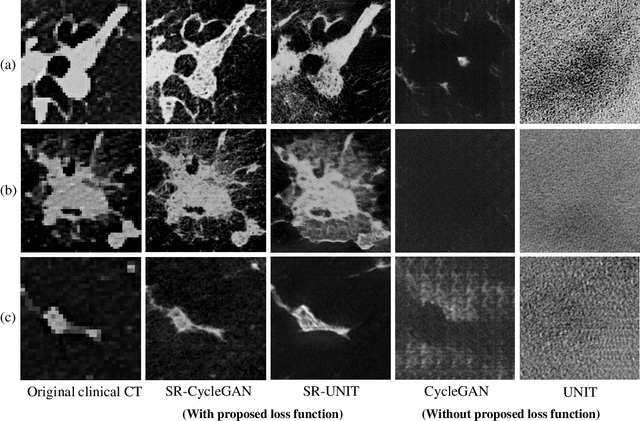
Abstract:This paper newly introduces multi-modality loss function for GAN-based super-resolution that can maintain image structure and intensity on unpaired training dataset of clinical CT and micro CT volumes. Precise non-invasive diagnosis of lung cancer mainly utilizes 3D multidetector computed-tomography (CT) data. On the other hand, we can take micro CT images of resected lung specimen in 50 micro meter or higher resolution. However, micro CT scanning cannot be applied to living human imaging. For obtaining highly detailed information such as cancer invasion area from pre-operative clinical CT volumes of lung cancer patients, super-resolution (SR) of clinical CT volumes to $\mu$CT level might be one of substitutive solutions. While most SR methods require paired low- and high-resolution images for training, it is infeasible to obtain precisely paired clinical CT and micro CT volumes. We aim to propose unpaired SR approaches for clincial CT using micro CT images based on unpaired image translation methods such as CycleGAN or UNIT. Since clinical CT and micro CT are very different in structure and intensity, direct application of GAN-based unpaired image translation methods in super-resolution tends to generate arbitrary images. Aiming to solve this problem, we propose new loss function called multi-modality loss function to maintain the similarity of input images and corresponding output images in super-resolution task. Experimental results demonstrated that the newly proposed loss function made CycleGAN and UNIT to successfully perform SR of clinical CT images of lung cancer patients into micro CT level resolution, while original CycleGAN and UNIT failed in super-resolution.
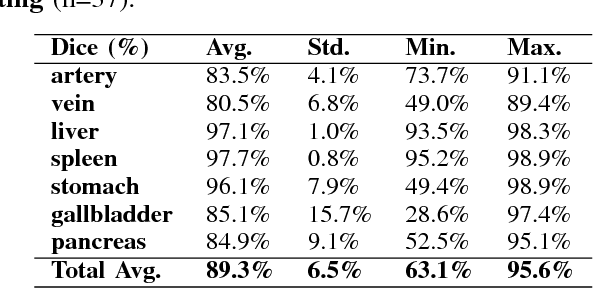
A multi-scale pyramid of 3D fully convolutional networks for abdominal multi-organ segmentation
Jun 06, 2018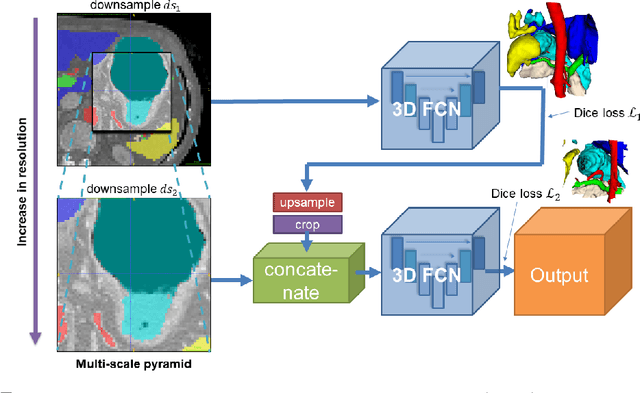
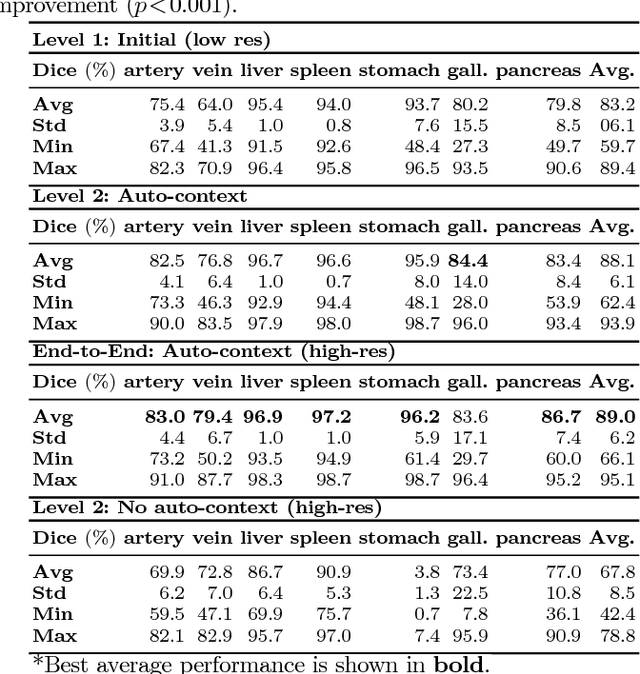
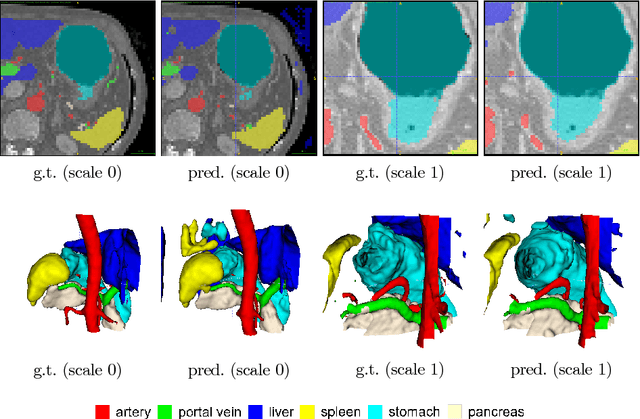
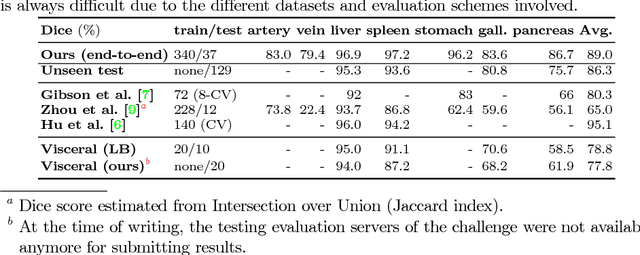
Abstract:Recent advances in deep learning, like 3D fully convolutional networks (FCNs), have improved the state-of-the-art in dense semantic segmentation of medical images. However, most network architectures require severely downsampling or cropping the images to meet the memory limitations of today's GPU cards while still considering enough context in the images for accurate segmentation. In this work, we propose a novel approach that utilizes auto-context to perform semantic segmentation at higher resolutions in a multi-scale pyramid of stacked 3D FCNs. We train and validate our models on a dataset of manually annotated abdominal organs and vessels from 377 clinical CT images used in gastric surgery, and achieve promising results with close to 90% Dice score on average. For additional evaluation, we perform separate testing on datasets from different sources and achieve competitive results, illustrating the robustness of the model and approach.
Unsupervised Segmentation of 3D Medical Images Based on Clustering and Deep Representation Learning
Apr 11, 2018Abstract:This paper presents a novel unsupervised segmentation method for 3D medical images. Convolutional neural networks (CNNs) have brought significant advances in image segmentation. However, most of the recent methods rely on supervised learning, which requires large amounts of manually annotated data. Thus, it is challenging for these methods to cope with the growing amount of medical images. This paper proposes a unified approach to unsupervised deep representation learning and clustering for segmentation. Our proposed method consists of two phases. In the first phase, we learn deep feature representations of training patches from a target image using joint unsupervised learning (JULE) that alternately clusters representations generated by a CNN and updates the CNN parameters using cluster labels as supervisory signals. We extend JULE to 3D medical images by utilizing 3D convolutions throughout the CNN architecture. In the second phase, we apply k-means to the deep representations from the trained CNN and then project cluster labels to the target image in order to obtain the fully segmented image. We evaluated our methods on three images of lung cancer specimens scanned with micro-computed tomography (micro-CT). The automatic segmentation of pathological regions in micro-CT could further contribute to the pathological examination process. Hence, we aim to automatically divide each image into the regions of invasive carcinoma, noninvasive carcinoma, and normal tissue. Our experiments show the potential abilities of unsupervised deep representation learning for medical image segmentation.
* This paper was presented at SPIE Medical Imaging 2018, Houston, TX, USA
Unsupervised Pathology Image Segmentation Using Representation Learning with Spherical K-means
Apr 11, 2018Abstract:This paper presents a novel method for unsupervised segmentation of pathology images. Staging of lung cancer is a major factor of prognosis. Measuring the maximum dimensions of the invasive component in a pathology images is an essential task. Therefore, image segmentation methods for visualizing the extent of invasive and noninvasive components on pathology images could support pathological examination. However, it is challenging for most of the recent segmentation methods that rely on supervised learning to cope with unlabeled pathology images. In this paper, we propose a unified approach to unsupervised representation learning and clustering for pathology image segmentation. Our method consists of two phases. In the first phase, we learn feature representations of training patches from a target image using the spherical k-means. The purpose of this phase is to obtain cluster centroids which could be used as filters for feature extraction. In the second phase, we apply conventional k-means to the representations extracted by the centroids and then project cluster labels to the target images. We evaluated our methods on pathology images of lung cancer specimen. Our experiments showed that the proposed method outperforms traditional k-means segmentation and the multithreshold Otsu method both quantitatively and qualitatively with an improved normalized mutual information (NMI) score of 0.626 compared to 0.168 and 0.167, respectively. Furthermore, we found that the centroids can be applied to the segmentation of other slices from the same sample.
* This paper was presented at SPIE Medical Imaging 2018, Houston, TX, USA
Deep learning and its application to medical image segmentation
Mar 23, 2018
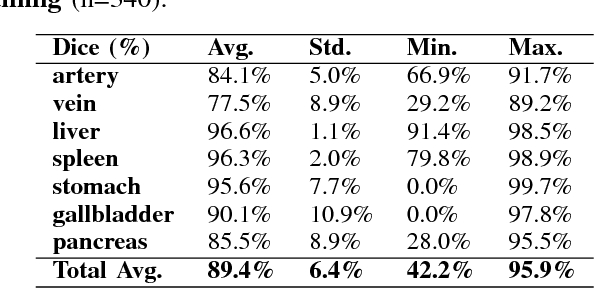
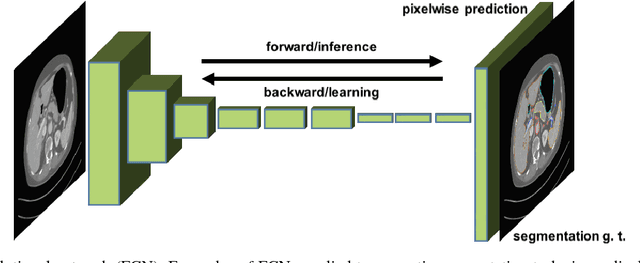
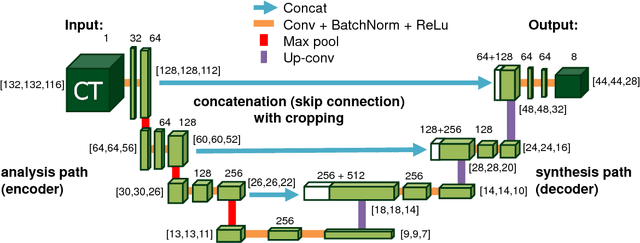
Abstract:One of the most common tasks in medical imaging is semantic segmentation. Achieving this segmentation automatically has been an active area of research, but the task has been proven very challenging due to the large variation of anatomy across different patients. However, recent advances in deep learning have made it possible to significantly improve the performance of image recognition and semantic segmentation methods in the field of computer vision. Due to the data driven approaches of hierarchical feature learning in deep learning frameworks, these advances can be translated to medical images without much difficulty. Several variations of deep convolutional neural networks have been successfully applied to medical images. Especially fully convolutional architectures have been proven efficient for segmentation of 3D medical images. In this article, we describe how to build a 3D fully convolutional network (FCN) that can process 3D images in order to produce automatic semantic segmentations. The model is trained and evaluated on a clinical computed tomography (CT) dataset and shows state-of-the-art performance in multi-organ segmentation.
* Accepted for publication in the journal of the Japanese Society of Medical Imaging Technology (JAMIT)
An application of cascaded 3D fully convolutional networks for medical image segmentation
Mar 20, 2018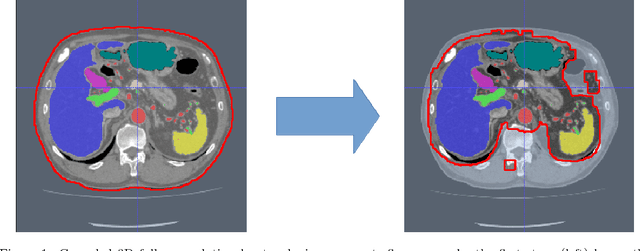
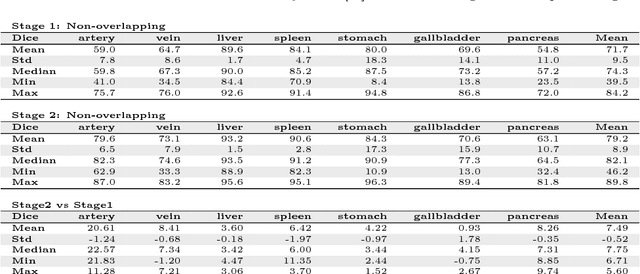
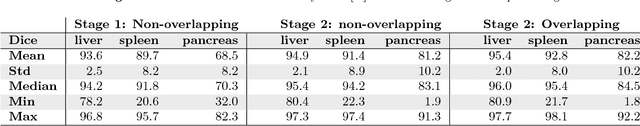
Abstract:Recent advances in 3D fully convolutional networks (FCN) have made it feasible to produce dense voxel-wise predictions of volumetric images. In this work, we show that a multi-class 3D FCN trained on manually labeled CT scans of several anatomical structures (ranging from the large organs to thin vessels) can achieve competitive segmentation results, while avoiding the need for handcrafting features or training class-specific models. To this end, we propose a two-stage, coarse-to-fine approach that will first use a 3D FCN to roughly define a candidate region, which will then be used as input to a second 3D FCN. This reduces the number of voxels the second FCN has to classify to ~10% and allows it to focus on more detailed segmentation of the organs and vessels. We utilize training and validation sets consisting of 331 clinical CT images and test our models on a completely unseen data collection acquired at a different hospital that includes 150 CT scans, targeting three anatomical organs (liver, spleen, and pancreas). In challenging organs such as the pancreas, our cascaded approach improves the mean Dice score from 68.5 to 82.2%, achieving the highest reported average score on this dataset. We compare with a 2D FCN method on a separate dataset of 240 CT scans with 18 classes and achieve a significantly higher performance in small organs and vessels. Furthermore, we explore fine-tuning our models to different datasets. Our experiments illustrate the promise and robustness of current 3D FCN based semantic segmentation of medical images, achieving state-of-the-art results. Our code and trained models are available for download: https://github.com/holgerroth/3Dunet_abdomen_cascade.
* Preprint accepted for publication in Computerized Medical Imaging and Graphics. Substantial extension of arXiv:1704.06382; Corrected references to figure numbers in this version
Towards dense volumetric pancreas segmentation in CT using 3D fully convolutional networks
Jan 19, 2018Abstract:Pancreas segmentation in computed tomography imaging has been historically difficult for automated methods because of the large shape and size variations between patients. In this work, we describe a custom-build 3D fully convolutional network (FCN) that can process a 3D image including the whole pancreas and produce an automatic segmentation. We investigate two variations of the 3D FCN architecture; one with concatenation and one with summation skip connections to the decoder part of the network. We evaluate our methods on a dataset from a clinical trial with gastric cancer patients, including 147 contrast enhanced abdominal CT scans acquired in the portal venous phase. Using the summation architecture, we achieve an average Dice score of 89.7 $\pm$ 3.8 (range [79.8, 94.8]) % in testing, achieving the new state-of-the-art performance in pancreas segmentation on this dataset.
* Accepted for oral presentation at SPIE Medical Imaging 2018, Houston, TX, USA Updated experiment in Fig. 4
On the influence of Dice loss function in multi-class organ segmentation of abdominal CT using 3D fully convolutional networks
Jan 18, 2018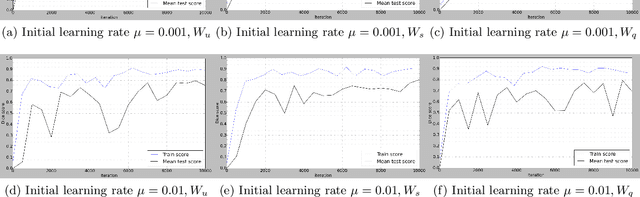

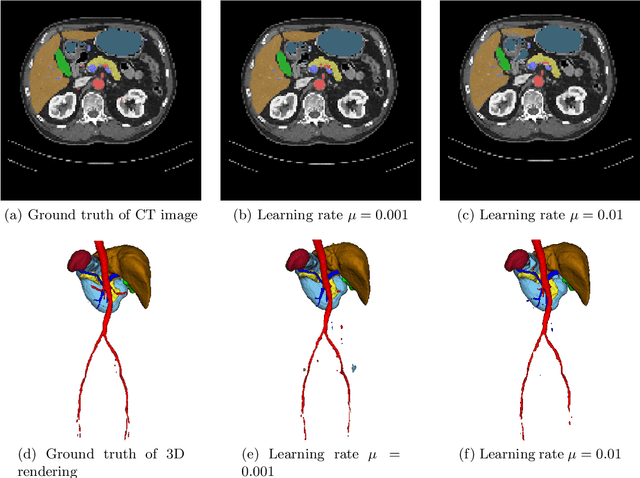
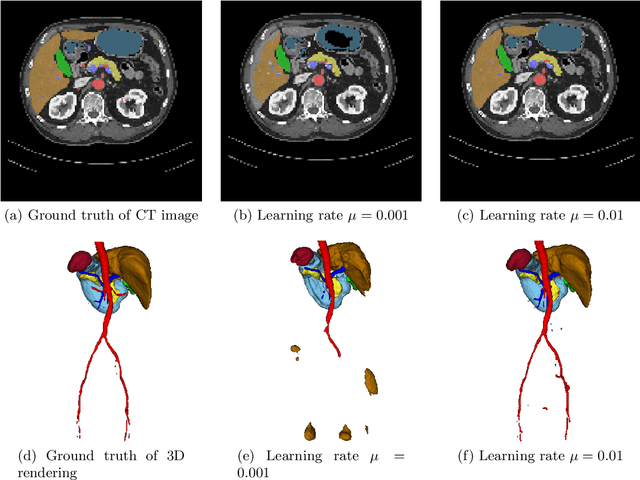
Abstract:Deep learning-based methods achieved impressive results for the segmentation of medical images. With the development of 3D fully convolutional networks (FCNs), it has become feasible to produce improved results for multi-organ segmentation of 3D computed tomography (CT) images. The results of multi-organ segmentation using deep learning-based methods not only depend on the choice of networks architecture, but also strongly rely on the choice of loss function. In this paper, we present a discussion on the influence of Dice-based loss functions for multi-class organ segmentation using a dataset of abdominal CT volumes. We investigated three different types of weighting the Dice loss functions based on class label frequencies (uniform, simple and square) and evaluate their influence on segmentation accuracies. Furthermore, we compared the influence of different initial learning rates. We achieved average Dice scores of 81.3%, 59.5% and 31.7% for uniform, simple and square types of weighting when the learning rate is 0.001, and 78.2%, 81.0% and 58.5% for each weighting when the learning rate is 0.01. Our experiments indicated a strong relationship between class balancing weights and initial learning rate in training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge