Natsuki Shimizu
Regression Forest-Based Atlas Localization and Direction Specific Atlas Generation for Pancreas Segmentation
May 07, 2020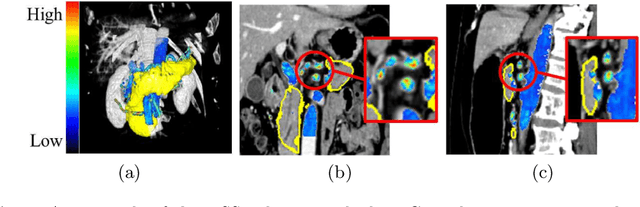
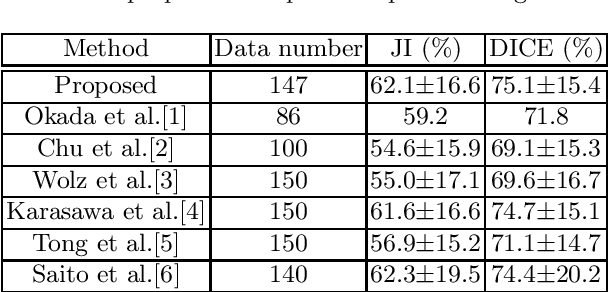


Abstract:This paper proposes a fully automated atlas-based pancreas segmentation method from CT volumes utilizing atlas localization by regression forest and atlas generation using blood vessel information. Previous probabilistic atlas-based pancreas segmentation methods cannot deal with spatial variations that are commonly found in the pancreas well. Also, shape variations are not represented by an averaged atlas. We propose a fully automated pancreas segmentation method that deals with two types of variations mentioned above. The position and size of the pancreas is estimated using a regression forest technique. After localization, a patient-specific probabilistic atlas is generated based on a new image similarity that reflects the blood vessel position and direction information around the pancreas. We segment it using the EM algorithm with the atlas as prior followed by the graph-cut. In evaluation results using 147 CT volumes, the Jaccard index and the Dice overlap of the proposed method were 62.1% and 75.1%, respectively. Although we automated all of the segmentation processes, segmentation results were superior to the other state-of-the-art methods in the Dice overlap.
* Accepted paper as a poster presentation at MICCAI 2016 (International Conference on Medical Image Computing and Computer-Assisted Intervention), Athens, Greece
3D FCN Feature Driven Regression Forest-Based Pancreas Localization and Segmentation
Jun 08, 2018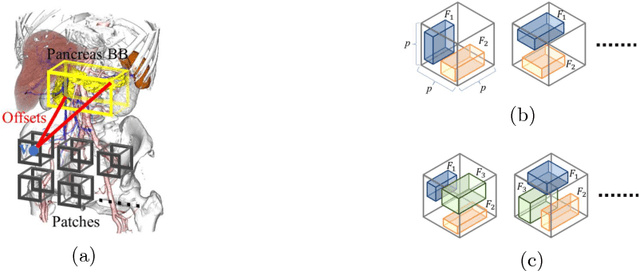

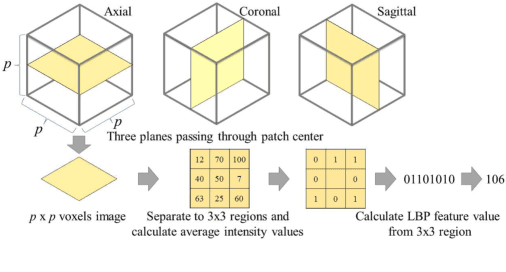
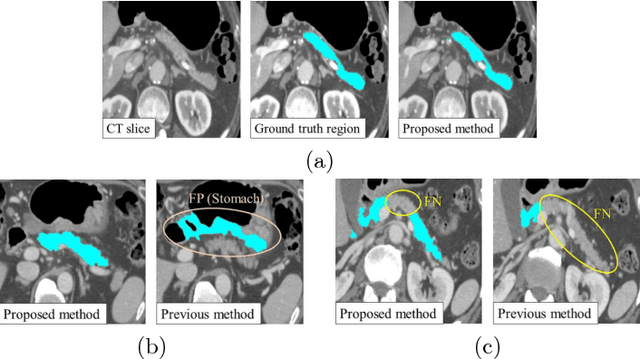
Abstract:This paper presents a fully automated atlas-based pancreas segmentation method from CT volumes utilizing 3D fully convolutional network (FCN) feature-based pancreas localization. Segmentation of the pancreas is difficult because it has larger inter-patient spatial variations than other organs. Previous pancreas segmentation methods failed to deal with such variations. We propose a fully automated pancreas segmentation method that contains novel localization and segmentation. Since the pancreas neighbors many other organs, its position and size are strongly related to the positions of the surrounding organs. We estimate the position and the size of the pancreas (localized) from global features by regression forests. As global features, we use intensity differences and 3D FCN deep learned features, which include automatically extracted essential features for segmentation. We chose 3D FCN features from a trained 3D U-Net, which is trained to perform multi-organ segmentation. The global features include both the pancreas and surrounding organ information. After localization, a patient-specific probabilistic atlas-based pancreas segmentation is performed. In evaluation results with 146 CT volumes, we achieved 60.6% of the Jaccard index and 73.9% of the Dice overlap.
* Presented in MICCAI 2017 workshop, DLMIA 2017 (Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support)
An application of cascaded 3D fully convolutional networks for medical image segmentation
Mar 20, 2018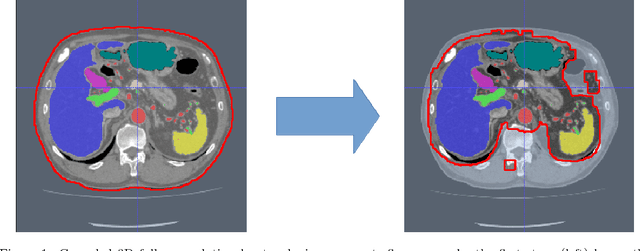
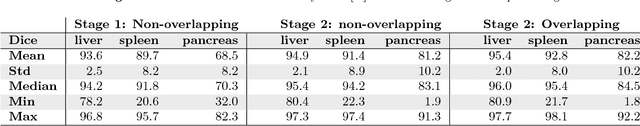
Abstract:Recent advances in 3D fully convolutional networks (FCN) have made it feasible to produce dense voxel-wise predictions of volumetric images. In this work, we show that a multi-class 3D FCN trained on manually labeled CT scans of several anatomical structures (ranging from the large organs to thin vessels) can achieve competitive segmentation results, while avoiding the need for handcrafting features or training class-specific models. To this end, we propose a two-stage, coarse-to-fine approach that will first use a 3D FCN to roughly define a candidate region, which will then be used as input to a second 3D FCN. This reduces the number of voxels the second FCN has to classify to ~10% and allows it to focus on more detailed segmentation of the organs and vessels. We utilize training and validation sets consisting of 331 clinical CT images and test our models on a completely unseen data collection acquired at a different hospital that includes 150 CT scans, targeting three anatomical organs (liver, spleen, and pancreas). In challenging organs such as the pancreas, our cascaded approach improves the mean Dice score from 68.5 to 82.2%, achieving the highest reported average score on this dataset. We compare with a 2D FCN method on a separate dataset of 240 CT scans with 18 classes and achieve a significantly higher performance in small organs and vessels. Furthermore, we explore fine-tuning our models to different datasets. Our experiments illustrate the promise and robustness of current 3D FCN based semantic segmentation of medical images, achieving state-of-the-art results. Our code and trained models are available for download: https://github.com/holgerroth/3Dunet_abdomen_cascade.
* Preprint accepted for publication in Computerized Medical Imaging and Graphics. Substantial extension of arXiv:1704.06382; Corrected references to figure numbers in this version
Towards dense volumetric pancreas segmentation in CT using 3D fully convolutional networks
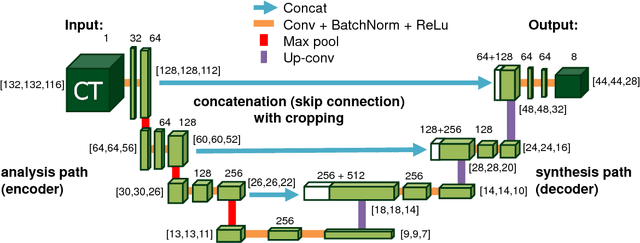
Jan 19, 2018Abstract:Pancreas segmentation in computed tomography imaging has been historically difficult for automated methods because of the large shape and size variations between patients. In this work, we describe a custom-build 3D fully convolutional network (FCN) that can process a 3D image including the whole pancreas and produce an automatic segmentation. We investigate two variations of the 3D FCN architecture; one with concatenation and one with summation skip connections to the decoder part of the network. We evaluate our methods on a dataset from a clinical trial with gastric cancer patients, including 147 contrast enhanced abdominal CT scans acquired in the portal venous phase. Using the summation architecture, we achieve an average Dice score of 89.7 $\pm$ 3.8 (range [79.8, 94.8]) % in testing, achieving the new state-of-the-art performance in pancreas segmentation on this dataset.
* Accepted for oral presentation at SPIE Medical Imaging 2018, Houston, TX, USA Updated experiment in Fig. 4
Hierarchical 3D fully convolutional networks for multi-organ segmentation
Apr 21, 2017



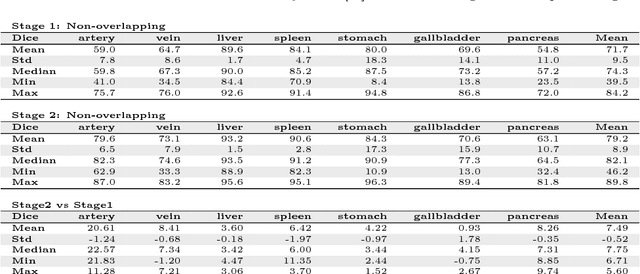
Abstract:Recent advances in 3D fully convolutional networks (FCN) have made it feasible to produce dense voxel-wise predictions of full volumetric images. In this work, we show that a multi-class 3D FCN trained on manually labeled CT scans of seven abdominal structures (artery, vein, liver, spleen, stomach, gallbladder, and pancreas) can achieve competitive segmentation results, while avoiding the need for handcrafting features or training organ-specific models. To this end, we propose a two-stage, coarse-to-fine approach that trains an FCN model to roughly delineate the organs of interest in the first stage (seeing $\sim$40% of the voxels within a simple, automatically generated binary mask of the patient's body). We then use these predictions of the first-stage FCN to define a candidate region that will be used to train a second FCN. This step reduces the number of voxels the FCN has to classify to $\sim$10% while maintaining a recall high of $>$99%. This second-stage FCN can now focus on more detailed segmentation of the organs. We respectively utilize training and validation sets consisting of 281 and 50 clinical CT images. Our hierarchical approach provides an improved Dice score of 7.5 percentage points per organ on average in our validation set. We furthermore test our models on a completely unseen data collection acquired at a different hospital that includes 150 CT scans with three anatomical labels (liver, spleen, and pancreas). In such challenging organs as the pancreas, our hierarchical approach improves the mean Dice score from 68.5 to 82.2%, achieving the highest reported average score on this dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge