Holger Roth
VILA-M3: Enhancing Vision-Language Models with Medical Expert Knowledge
Nov 19, 2024



Abstract:Generalist vision language models (VLMs) have made significant strides in computer vision, but they fall short in specialized fields like healthcare, where expert knowledge is essential. In traditional computer vision tasks, creative or approximate answers may be acceptable, but in healthcare, precision is paramount.Current large multimodal models like Gemini and GPT-4o are insufficient for medical tasks due to their reliance on memorized internet knowledge rather than the nuanced expertise required in healthcare. VLMs are usually trained in three stages: vision pre-training, vision-language pre-training, and instruction fine-tuning (IFT). IFT has been typically applied using a mixture of generic and healthcare data. In contrast, we propose that for medical VLMs, a fourth stage of specialized IFT is necessary, which focuses on medical data and includes information from domain expert models. Domain expert models developed for medical use are crucial because they are specifically trained for certain clinical tasks, e.g. to detect tumors and classify abnormalities through segmentation and classification, which learn fine-grained features of medical data$-$features that are often too intricate for a VLM to capture effectively especially in radiology. This paper introduces a new framework, VILA-M3, for medical VLMs that utilizes domain knowledge via expert models. Through our experiments, we show an improved state-of-the-art (SOTA) performance with an average improvement of ~9% over the prior SOTA model Med-Gemini and ~6% over models trained on the specific tasks. Our approach emphasizes the importance of domain expertise in creating precise, reliable VLMs for medical applications.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
HoloHisto: End-to-end Gigapixel WSI Segmentation with 4K Resolution Sequential Tokenization
Jul 03, 2024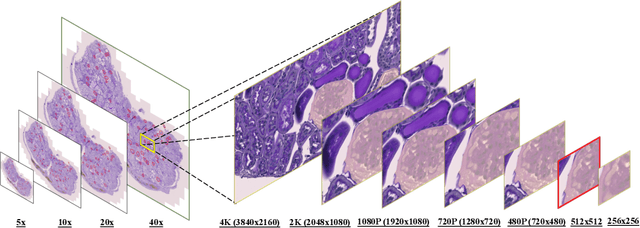
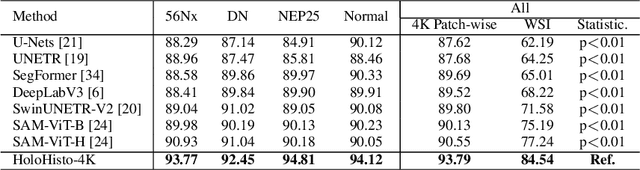
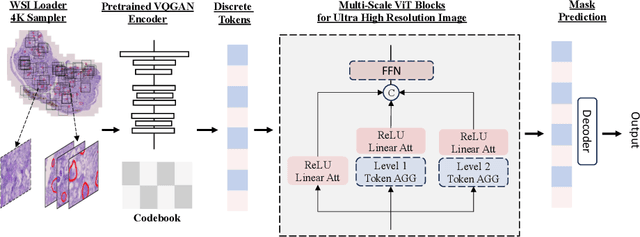
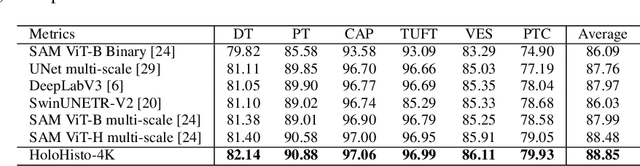
Abstract:In digital pathology, the traditional method for deep learning-based image segmentation typically involves a two-stage process: initially segmenting high-resolution whole slide images (WSI) into smaller patches (e.g., 256x256, 512x512, 1024x1024) and subsequently reconstructing them to their original scale. This method often struggles to capture the complex details and vast scope of WSIs. In this paper, we propose the holistic histopathology (HoloHisto) segmentation method to achieve end-to-end segmentation on gigapixel WSIs, whose maximum resolution is above 80,000$\times$70,000 pixels. HoloHisto fundamentally shifts the paradigm of WSI segmentation to an end-to-end learning fashion with 1) a large (4K) resolution base patch for elevated visual information inclusion and efficient processing, and 2) a novel sequential tokenization mechanism to properly model the contextual relationships and efficiently model the rich information from the 4K input. To our best knowledge, HoloHisto presents the first holistic approach for gigapixel resolution WSI segmentation, supporting direct I/O of complete WSI and their corresponding gigapixel masks. Under the HoloHisto platform, we unveil a random 4K sampler that transcends ultra-high resolution, delivering 31 and 10 times more pixels than standard 2D and 3D patches, respectively, for advancing computational capabilities. To facilitate efficient 4K resolution dense prediction, we leverage sequential tokenization, utilizing a pre-trained image tokenizer to group image features into a discrete token grid. To assess the performance, our team curated a new kidney pathology image segmentation (KPIs) dataset with WSI-level glomeruli segmentation from whole mouse kidneys. From the results, HoloHisto-4K delivers remarkable performance gains over previous state-of-the-art models.
UNetFormer: A Unified Vision Transformer Model and Pre-Training Framework for 3D Medical Image Segmentation
Apr 05, 2022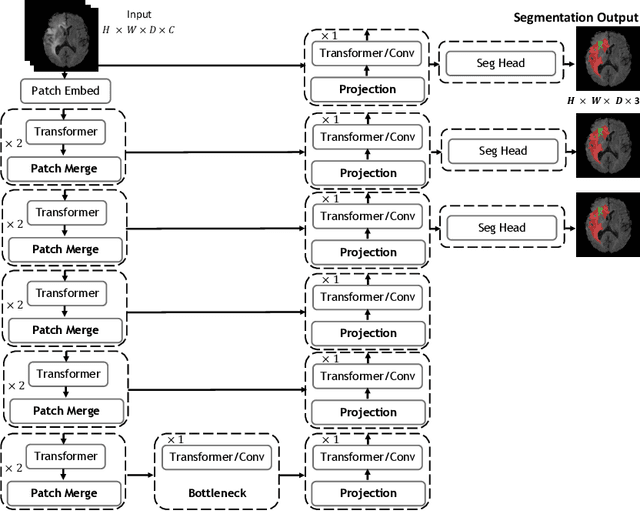
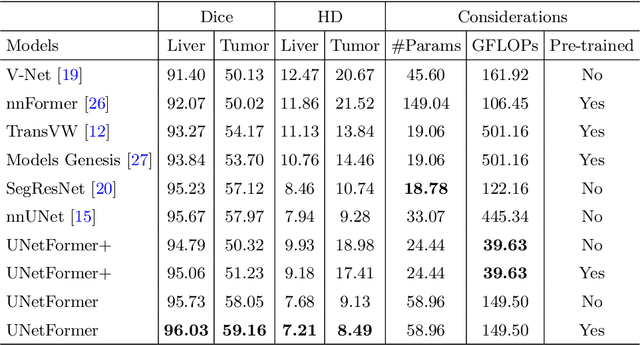
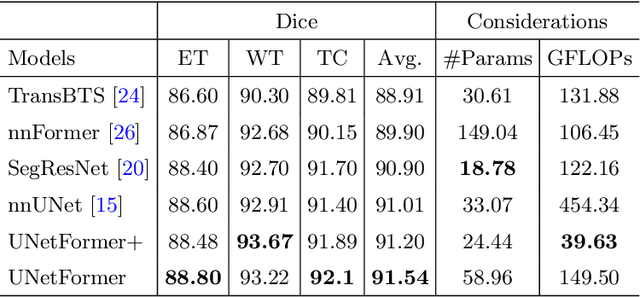
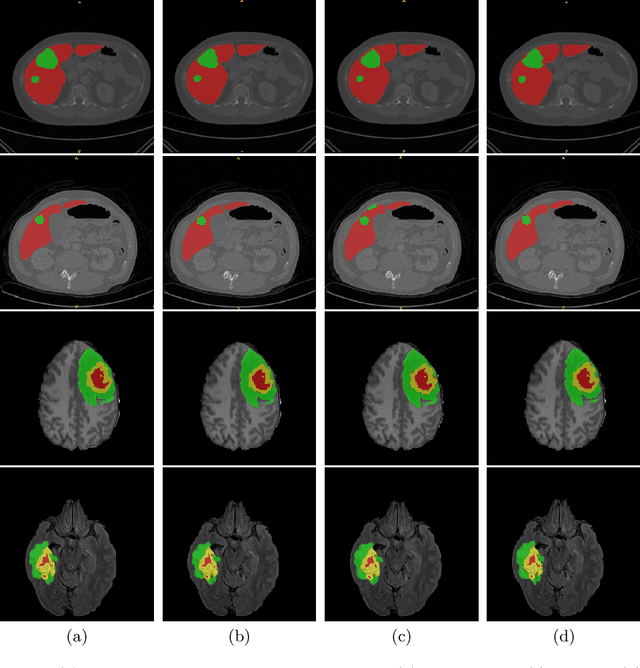
Abstract:Vision Transformers (ViT)s have recently become popular due to their outstanding modeling capabilities, in particular for capturing long-range information, and scalability to dataset and model sizes which has led to state-of-the-art performance in various computer vision and medical image analysis tasks. In this work, we introduce a unified framework consisting of two architectures, dubbed UNetFormer, with a 3D Swin Transformer-based encoder and Convolutional Neural Network (CNN) and transformer-based decoders. In the proposed model, the encoder is linked to the decoder via skip connections at five different resolutions with deep supervision. The design of proposed architecture allows for meeting a wide range of trade-off requirements between accuracy and computational cost. In addition, we present a methodology for self-supervised pre-training of the encoder backbone via learning to predict randomly masked volumetric tokens using contextual information of visible tokens. We pre-train our framework on a cohort of $5050$ CT images, gathered from publicly available CT datasets, and present a systematic investigation of various components such as masking ratio and patch size that affect the representation learning capability and performance of downstream tasks. We validate the effectiveness of our pre-training approach by fine-tuning and testing our model on liver and liver tumor segmentation task using the Medical Segmentation Decathlon (MSD) dataset and achieve state-of-the-art performance in terms of various segmentation metrics. To demonstrate its generalizability, we train and test the model on BraTS 21 dataset for brain tumor segmentation using MRI images and outperform other methods in terms of Dice score. Code: https://github.com/Project-MONAI/research-contributions
GradViT: Gradient Inversion of Vision Transformers
Mar 28, 2022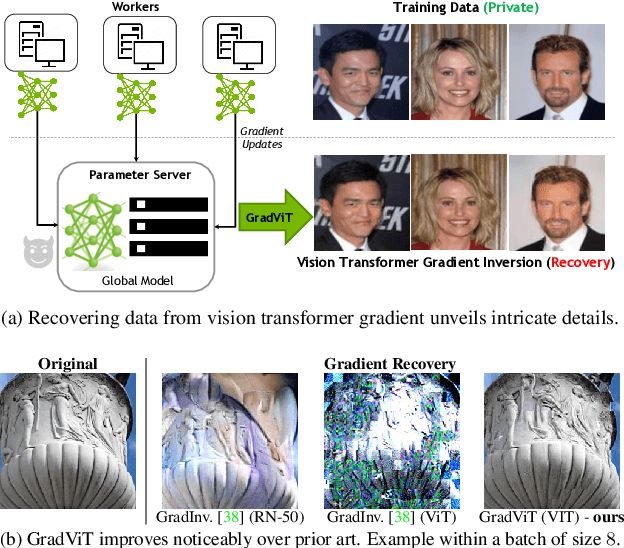
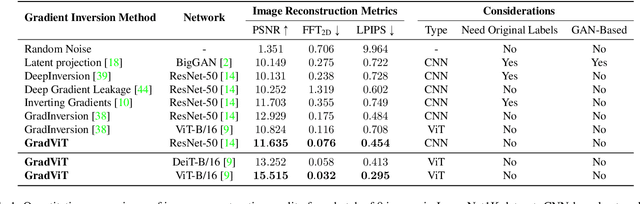
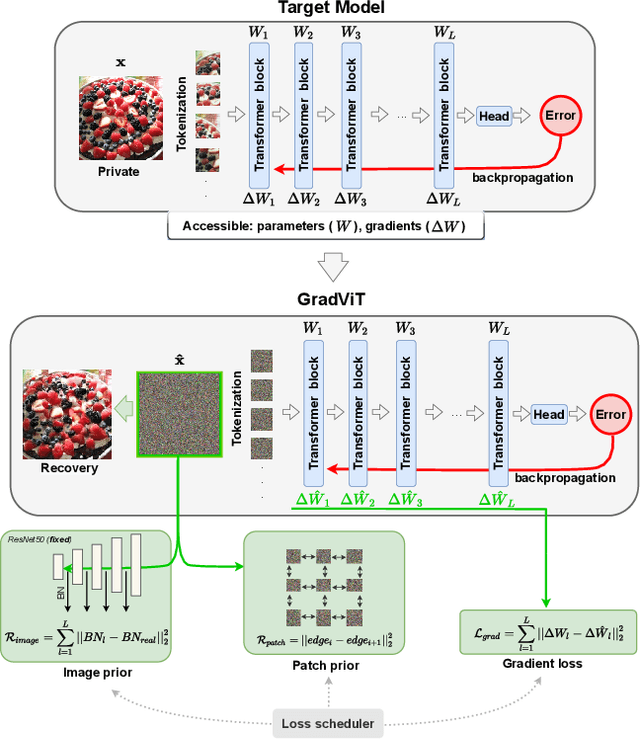
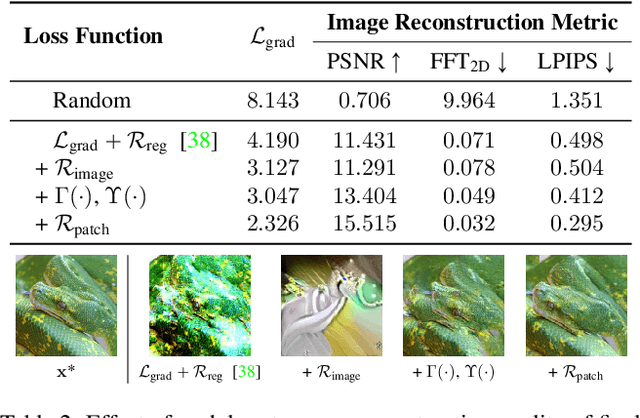
Abstract:In this work we demonstrate the vulnerability of vision transformers (ViTs) to gradient-based inversion attacks. During this attack, the original data batch is reconstructed given model weights and the corresponding gradients. We introduce a method, named GradViT, that optimizes random noise into naturally looking images via an iterative process. The optimization objective consists of (i) a loss on matching the gradients, (ii) image prior in the form of distance to batch-normalization statistics of a pretrained CNN model, and (iii) a total variation regularization on patches to guide correct recovery locations. We propose a unique loss scheduling function to overcome local minima during optimization. We evaluate GadViT on ImageNet1K and MS-Celeb-1M datasets, and observe unprecedentedly high fidelity and closeness to the original (hidden) data. During the analysis we find that vision transformers are significantly more vulnerable than previously studied CNNs due to the presence of the attention mechanism. Our method demonstrates new state-of-the-art results for gradient inversion in both qualitative and quantitative metrics. Project page at https://gradvit.github.io/.
Closing the Generalization Gap of Cross-silo Federated Medical Image Segmentation
Mar 18, 2022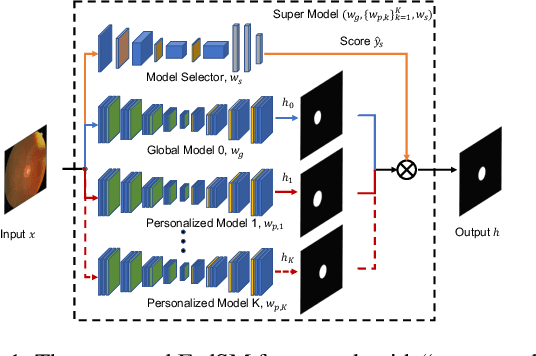
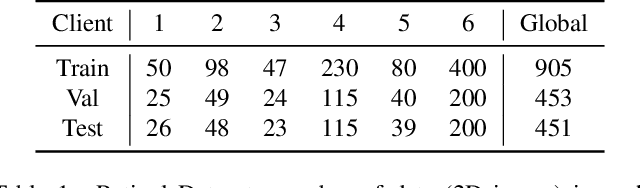
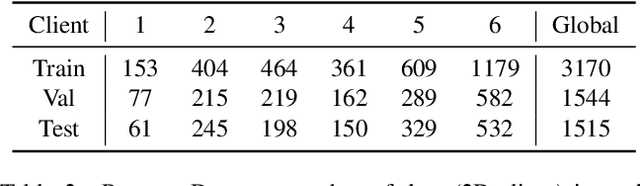
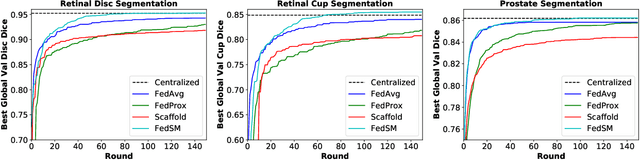
Abstract:Cross-silo federated learning (FL) has attracted much attention in medical imaging analysis with deep learning in recent years as it can resolve the critical issues of insufficient data, data privacy, and training efficiency. However, there can be a generalization gap between the model trained from FL and the one from centralized training. This important issue comes from the non-iid data distribution of the local data in the participating clients and is well-known as client drift. In this work, we propose a novel training framework FedSM to avoid the client drift issue and successfully close the generalization gap compared with the centralized training for medical image segmentation tasks for the first time. We also propose a novel personalized FL objective formulation and a new method SoftPull to solve it in our proposed framework FedSM. We conduct rigorous theoretical analysis to guarantee its convergence for optimizing the non-convex smooth objective function. Real-world medical image segmentation experiments using deep FL validate the motivations and effectiveness of our proposed method.
Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images
Jan 04, 2022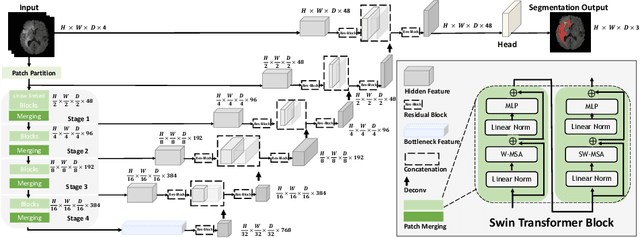

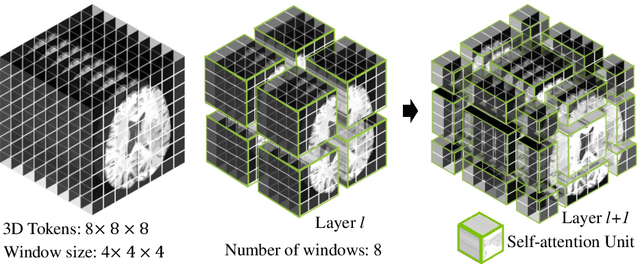
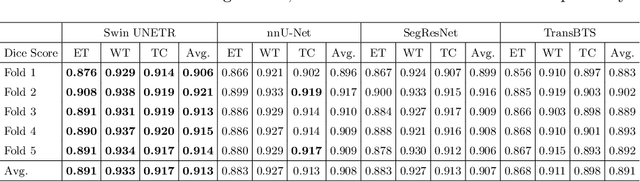
Abstract:Semantic segmentation of brain tumors is a fundamental medical image analysis task involving multiple MRI imaging modalities that can assist clinicians in diagnosing the patient and successively studying the progression of the malignant entity. In recent years, Fully Convolutional Neural Networks (FCNNs) approaches have become the de facto standard for 3D medical image segmentation. The popular "U-shaped" network architecture has achieved state-of-the-art performance benchmarks on different 2D and 3D semantic segmentation tasks and across various imaging modalities. However, due to the limited kernel size of convolution layers in FCNNs, their performance of modeling long-range information is sub-optimal, and this can lead to deficiencies in the segmentation of tumors with variable sizes. On the other hand, transformer models have demonstrated excellent capabilities in capturing such long-range information in multiple domains, including natural language processing and computer vision. Inspired by the success of vision transformers and their variants, we propose a novel segmentation model termed Swin UNEt TRansformers (Swin UNETR). Specifically, the task of 3D brain tumor semantic segmentation is reformulated as a sequence to sequence prediction problem wherein multi-modal input data is projected into a 1D sequence of embedding and used as an input to a hierarchical Swin transformer as the encoder. The swin transformer encoder extracts features at five different resolutions by utilizing shifted windows for computing self-attention and is connected to an FCNN-based decoder at each resolution via skip connections. We have participated in BraTS 2021 segmentation challenge, and our proposed model ranks among the top-performing approaches in the validation phase. Code: https://monai.io/research/swin-unetr
Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis
Nov 29, 2021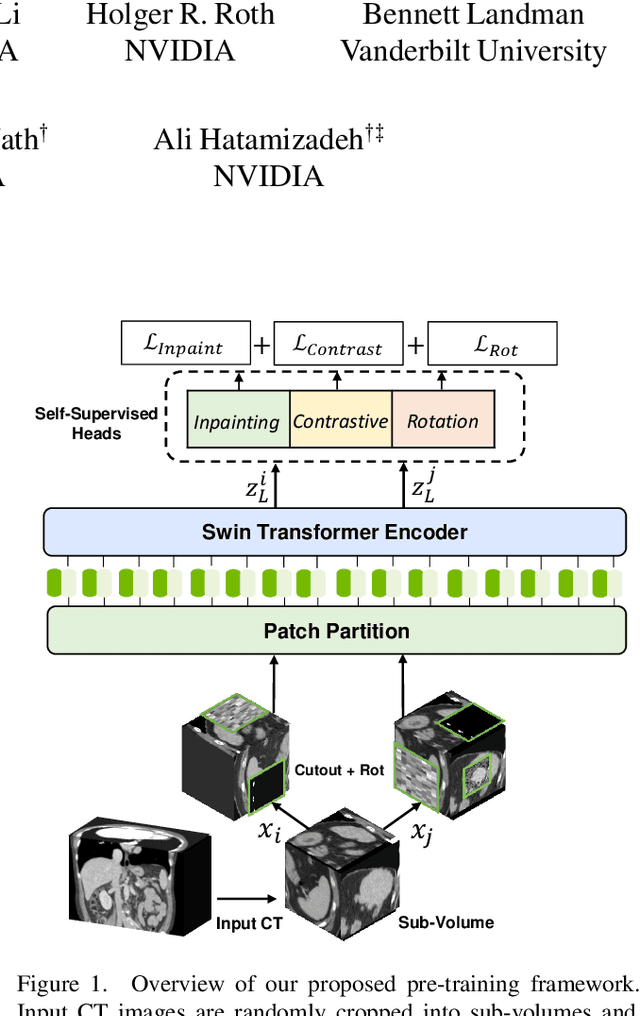
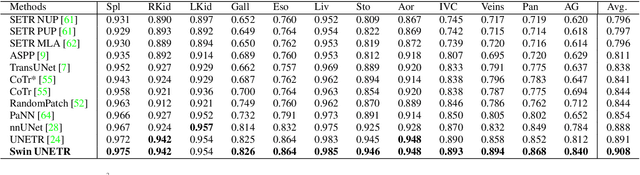
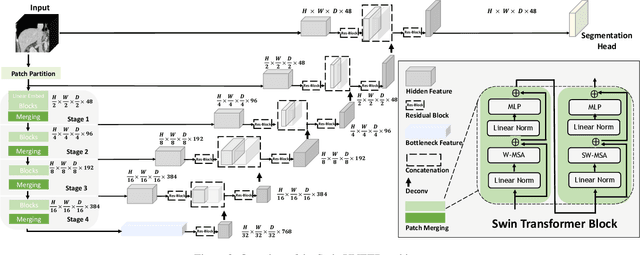
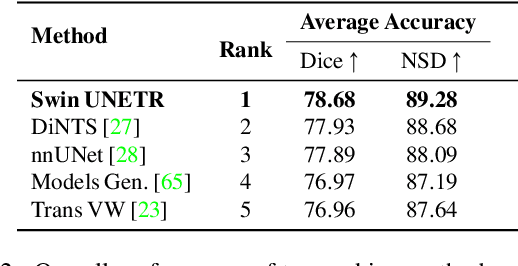
Abstract:Vision Transformers (ViT)s have shown great performance in self-supervised learning of global and local representations that can be transferred to downstream applications. Inspired by these results, we introduce a novel self-supervised learning framework with tailored proxy tasks for medical image analysis. Specifically, we propose: (i) a new 3D transformer-based model, dubbed Swin UNEt TRansformers (Swin UNETR), with a hierarchical encoder for self-supervised pre-training; (ii) tailored proxy tasks for learning the underlying pattern of human anatomy. We demonstrate successful pre-training of the proposed model on 5,050 publicly available computed tomography (CT) images from various body organs. The effectiveness of our approach is validated by fine-tuning the pre-trained models on the Beyond the Cranial Vault (BTCV) Segmentation Challenge with 13 abdominal organs and segmentation tasks from the Medical Segmentation Decathlon (MSD) dataset. Our model is currently the state-of-the-art (i.e. ranked 1st) on the public test leaderboards of both MSD and BTCV datasets. Code: https://monai.io/research/swin-unetr
Accounting for Dependencies in Deep Learning Based Multiple Instance Learning for Whole Slide Imaging
Nov 01, 2021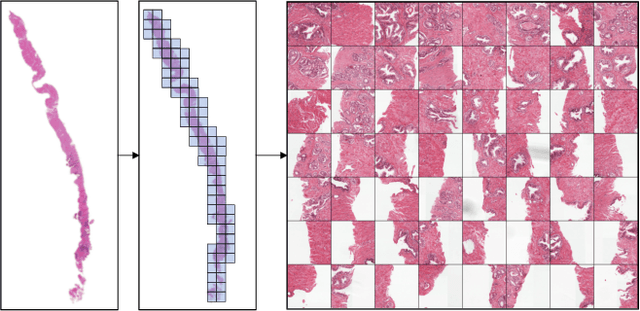
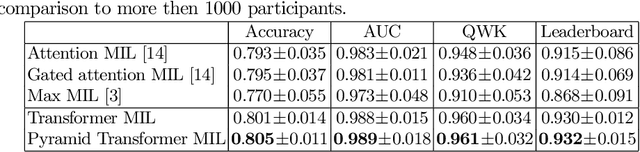
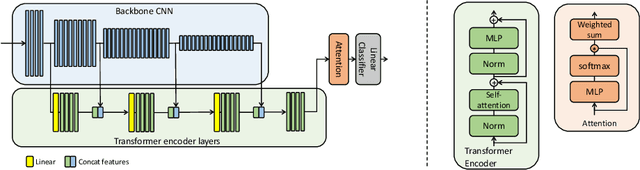
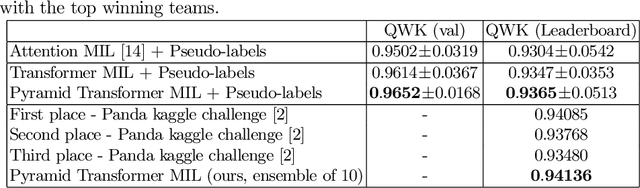
Abstract:Multiple instance learning (MIL) is a key algorithm for classification of whole slide images (WSI). Histology WSIs can have billions of pixels, which create enormous computational and annotation challenges. Typically, such images are divided into a set of patches (a bag of instances), where only bag-level class labels are provided. Deep learning based MIL methods calculate instance features using convolutional neural network (CNN). Our proposed approach is also deep learning based, with the following two contributions: Firstly, we propose to explicitly account for dependencies between instances during training by embedding self-attention Transformer blocks to capture dependencies between instances. For example, a tumor grade may depend on the presence of several particular patterns at different locations in WSI, which requires to account for dependencies between patches. Secondly, we propose an instance-wise loss function based on instance pseudo-labels. We compare the proposed algorithm to multiple baseline methods, evaluate it on the PANDA challenge dataset, the largest publicly available WSI dataset with over 11K images, and demonstrate state-of-the-art results.
The Power of Proxy Data and Proxy Networks for Hyper-Parameter Optimization in Medical Image Segmentation
Jul 12, 2021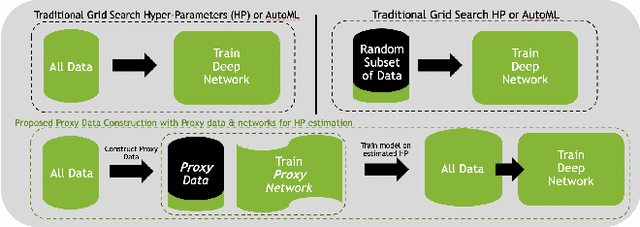
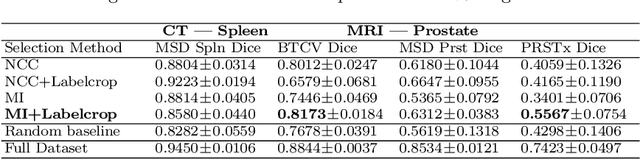
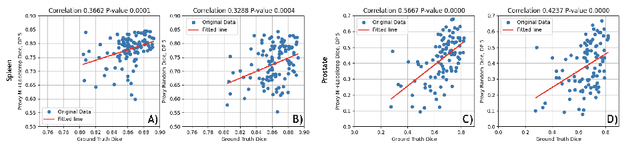
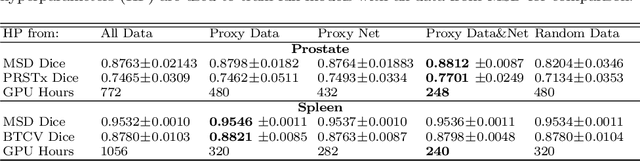
Abstract:Deep learning models for medical image segmentation are primarily data-driven. Models trained with more data lead to improved performance and generalizability. However, training is a computationally expensive process because multiple hyper-parameters need to be tested to find the optimal setting for best performance. In this work, we focus on accelerating the estimation of hyper-parameters by proposing two novel methodologies: proxy data and proxy networks. Both can be useful for estimating hyper-parameters more efficiently. We test the proposed techniques on CT and MR imaging modalities using well-known public datasets. In both cases using one dataset for building proxy data and another data source for external evaluation. For CT, the approach is tested on spleen segmentation with two datasets. The first dataset is from the medical segmentation decathlon (MSD), where the proxy data is constructed, the secondary dataset is utilized as an external validation dataset. Similarly, for MR, the approach is evaluated on prostate segmentation where the first dataset is from MSD and the second dataset is PROSTATEx. First, we show higher correlation to using full data for training when testing on the external validation set using smaller proxy data than a random selection of the proxy data. Second, we show that a high correlation exists for proxy networks when compared with the full network on validation Dice score. Third, we show that the proposed approach of utilizing a proxy network can speed up an AutoML framework for hyper-parameter search by 3.3x, and by 4.4x if proxy data and proxy network are utilized together.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge