Yufan He
SurgWorld: Learning Surgical Robot Policies from Videos via World Modeling
Dec 30, 2025Abstract:Data scarcity remains a fundamental barrier to achieving fully autonomous surgical robots. While large scale vision language action (VLA) models have shown impressive generalization in household and industrial manipulation by leveraging paired video action data from diverse domains, surgical robotics suffers from the paucity of datasets that include both visual observations and accurate robot kinematics. In contrast, vast corpora of surgical videos exist, but they lack corresponding action labels, preventing direct application of imitation learning or VLA training. In this work, we aim to alleviate this problem by learning policy models from SurgWorld, a world model designed for surgical physical AI. We curated the Surgical Action Text Alignment (SATA) dataset with detailed action description specifically for surgical robots. Then we built SurgeWorld based on the most advanced physical AI world model and SATA. It's able to generate diverse, generalizable and realistic surgery videos. We are also the first to use an inverse dynamics model to infer pseudokinematics from synthetic surgical videos, producing synthetic paired video action data. We demonstrate that a surgical VLA policy trained with these augmented data significantly outperforms models trained only on real demonstrations on a real surgical robot platform. Our approach offers a scalable path toward autonomous surgical skill acquisition by leveraging the abundance of unlabeled surgical video and generative world modeling, thus opening the door to generalizable and data efficient surgical robot policies.
Text2CT: Towards 3D CT Volume Generation from Free-text Descriptions Using Diffusion Model
May 07, 2025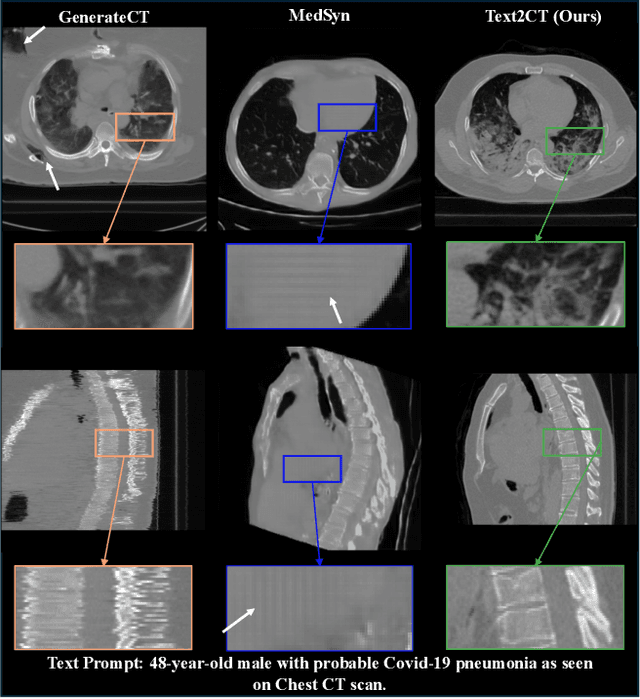
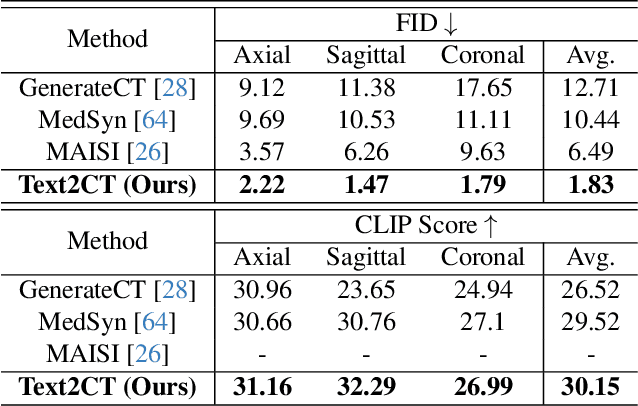
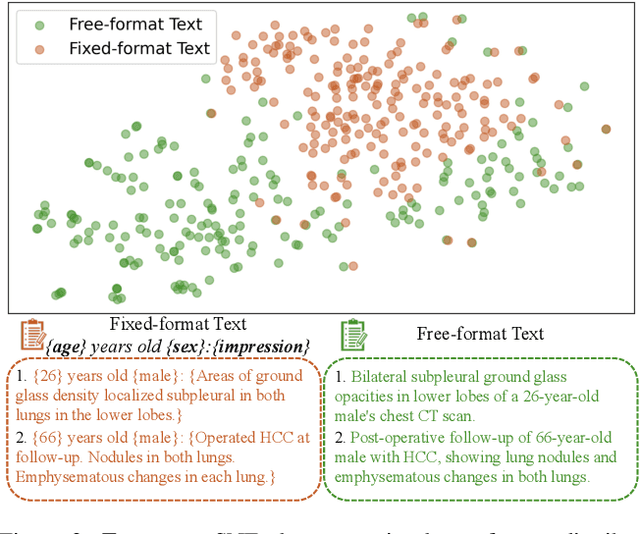
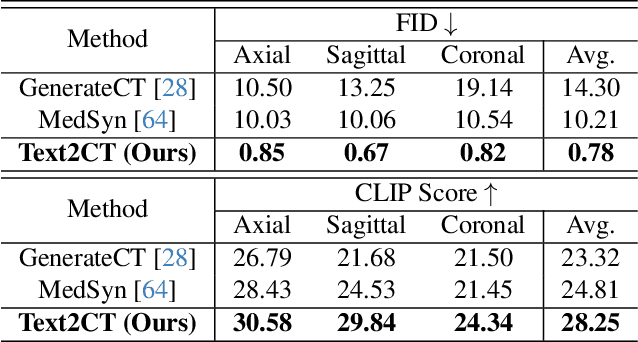
Abstract:Generating 3D CT volumes from descriptive free-text inputs presents a transformative opportunity in diagnostics and research. In this paper, we introduce Text2CT, a novel approach for synthesizing 3D CT volumes from textual descriptions using the diffusion model. Unlike previous methods that rely on fixed-format text input, Text2CT employs a novel prompt formulation that enables generation from diverse, free-text descriptions. The proposed framework encodes medical text into latent representations and decodes them into high-resolution 3D CT scans, effectively bridging the gap between semantic text inputs and detailed volumetric representations in a unified 3D framework. Our method demonstrates superior performance in preserving anatomical fidelity and capturing intricate structures as described in the input text. Extensive evaluations show that our approach achieves state-of-the-art results, offering promising potential applications in diagnostics, and data augmentation.
Unsupervised learning of spatially varying regularization for diffeomorphic image registration
Dec 23, 2024Abstract:Spatially varying regularization accommodates the deformation variations that may be necessary for different anatomical regions during deformable image registration. Historically, optimization-based registration models have harnessed spatially varying regularization to address anatomical subtleties. However, most modern deep learning-based models tend to gravitate towards spatially invariant regularization, wherein a homogenous regularization strength is applied across the entire image, potentially disregarding localized variations. In this paper, we propose a hierarchical probabilistic model that integrates a prior distribution on the deformation regularization strength, enabling the end-to-end learning of a spatially varying deformation regularizer directly from the data. The proposed method is straightforward to implement and easily integrates with various registration network architectures. Additionally, automatic tuning of hyperparameters is achieved through Bayesian optimization, allowing efficient identification of optimal hyperparameters for any given registration task. Comprehensive evaluations on publicly available datasets demonstrate that the proposed method significantly improves registration performance and enhances the interpretability of deep learning-based registration, all while maintaining smooth deformations.
VILA-M3: Enhancing Vision-Language Models with Medical Expert Knowledge
Nov 19, 2024



Abstract:Generalist vision language models (VLMs) have made significant strides in computer vision, but they fall short in specialized fields like healthcare, where expert knowledge is essential. In traditional computer vision tasks, creative or approximate answers may be acceptable, but in healthcare, precision is paramount.Current large multimodal models like Gemini and GPT-4o are insufficient for medical tasks due to their reliance on memorized internet knowledge rather than the nuanced expertise required in healthcare. VLMs are usually trained in three stages: vision pre-training, vision-language pre-training, and instruction fine-tuning (IFT). IFT has been typically applied using a mixture of generic and healthcare data. In contrast, we propose that for medical VLMs, a fourth stage of specialized IFT is necessary, which focuses on medical data and includes information from domain expert models. Domain expert models developed for medical use are crucial because they are specifically trained for certain clinical tasks, e.g. to detect tumors and classify abnormalities through segmentation and classification, which learn fine-grained features of medical data$-$features that are often too intricate for a VLM to capture effectively especially in radiology. This paper introduces a new framework, VILA-M3, for medical VLMs that utilizes domain knowledge via expert models. Through our experiments, we show an improved state-of-the-art (SOTA) performance with an average improvement of ~9% over the prior SOTA model Med-Gemini and ~6% over models trained on the specific tasks. Our approach emphasizes the importance of domain expertise in creating precise, reliable VLMs for medical applications.
A Short Review and Evaluation of SAM2's Performance in 3D CT Image Segmentation
Aug 20, 2024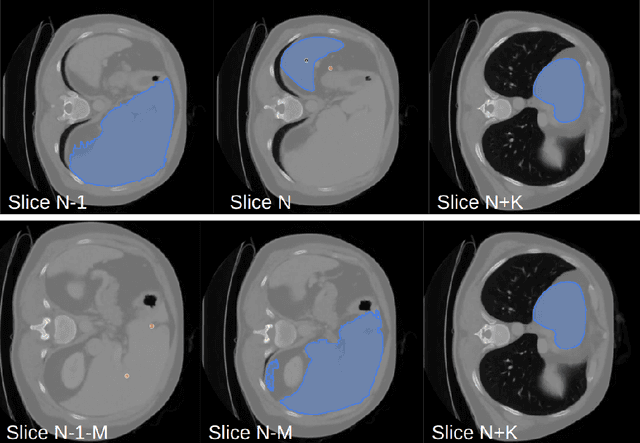
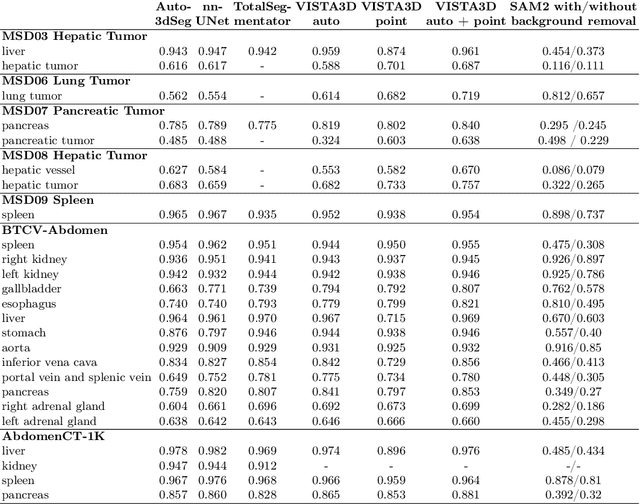
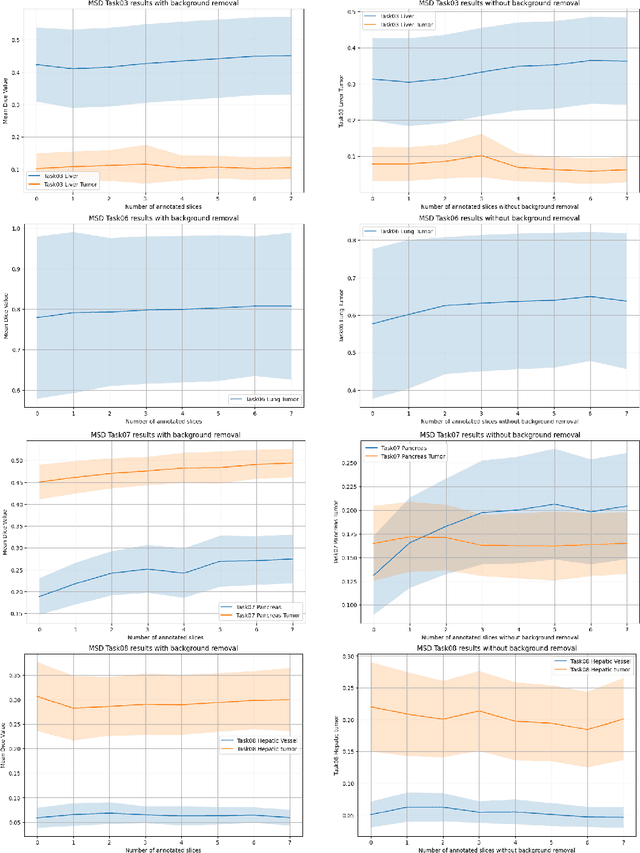
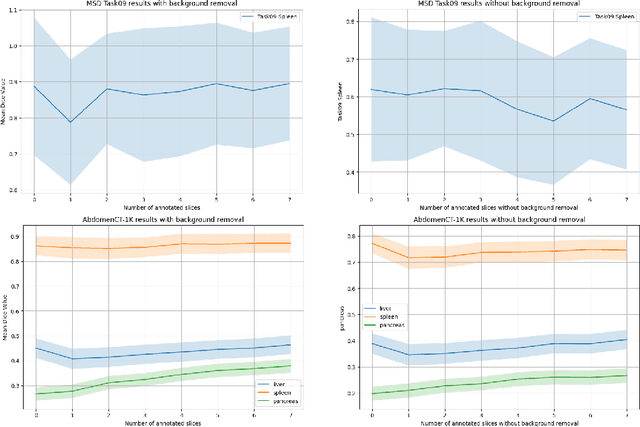
Abstract:Since the release of Segment Anything 2 (SAM2), the medical imaging community has been actively evaluating its performance for 3D medical image segmentation. However, different studies have employed varying evaluation pipelines, resulting in conflicting outcomes that obscure a clear understanding of SAM2's capabilities and potential applications. We shortly review existing benchmarks and point out that the SAM2 paper clearly outlines a zero-shot evaluation pipeline, which simulates user clicks iteratively for up to eight iterations. We reproduced this interactive annotation simulation on 3D CT datasets and provided the results and code~\url{https://github.com/Project-MONAI/VISTA}. Our findings reveal that directly applying SAM2 on 3D medical imaging in a zero-shot manner is far from satisfactory. It is prone to generating false positives when foreground objects disappear, and annotating more slices cannot fully offset this tendency. For smaller single-connected objects like kidney and aorta, SAM2 performs reasonably well but for most organs it is still far behind state-of-the-art 3D annotation methods. More research and innovation are needed for 3D medical imaging community to use SAM2 correctly.
HoloHisto: End-to-end Gigapixel WSI Segmentation with 4K Resolution Sequential Tokenization
Jul 03, 2024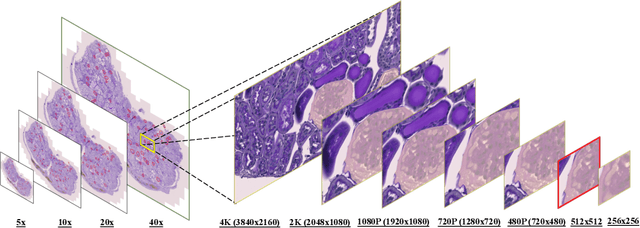
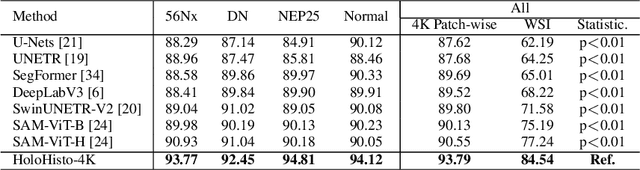
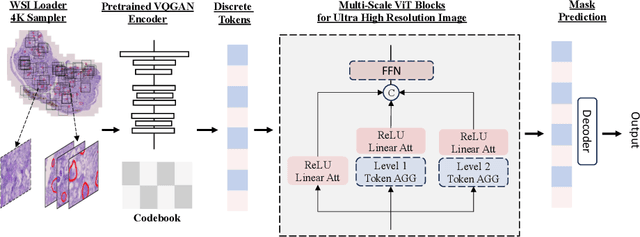
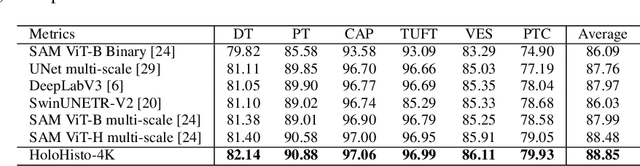
Abstract:In digital pathology, the traditional method for deep learning-based image segmentation typically involves a two-stage process: initially segmenting high-resolution whole slide images (WSI) into smaller patches (e.g., 256x256, 512x512, 1024x1024) and subsequently reconstructing them to their original scale. This method often struggles to capture the complex details and vast scope of WSIs. In this paper, we propose the holistic histopathology (HoloHisto) segmentation method to achieve end-to-end segmentation on gigapixel WSIs, whose maximum resolution is above 80,000$\times$70,000 pixels. HoloHisto fundamentally shifts the paradigm of WSI segmentation to an end-to-end learning fashion with 1) a large (4K) resolution base patch for elevated visual information inclusion and efficient processing, and 2) a novel sequential tokenization mechanism to properly model the contextual relationships and efficiently model the rich information from the 4K input. To our best knowledge, HoloHisto presents the first holistic approach for gigapixel resolution WSI segmentation, supporting direct I/O of complete WSI and their corresponding gigapixel masks. Under the HoloHisto platform, we unveil a random 4K sampler that transcends ultra-high resolution, delivering 31 and 10 times more pixels than standard 2D and 3D patches, respectively, for advancing computational capabilities. To facilitate efficient 4K resolution dense prediction, we leverage sequential tokenization, utilizing a pre-trained image tokenizer to group image features into a discrete token grid. To assess the performance, our team curated a new kidney pathology image segmentation (KPIs) dataset with WSI-level glomeruli segmentation from whole mouse kidneys. From the results, HoloHisto-4K delivers remarkable performance gains over previous state-of-the-art models.
VISTA3D: Versatile Imaging SegmenTation and Annotation model for 3D Computed Tomography
Jun 07, 2024Abstract:Segmentation foundation models have attracted great interest, however, none of them are adequate enough for the use cases in 3D computed tomography scans (CT) images. Existing works finetune on medical images with 2D foundation models trained on natural images, but interactive segmentation, especially in 2D, is too time-consuming for 3D scans and less useful for large cohort analysis. Models that can perform out-of-the-box automatic segmentation are more desirable. However, the model trained in this way lacks the ability to perform segmentation on unseen objects like novel tumors. Thus for 3D medical image analysis, an ideal segmentation solution might expect two features: accurate out-of-the-box performance covering major organ classes, and effective adaptation or zero-shot ability to novel structures. In this paper, we discuss what features a 3D CT segmentation foundation model should have, and introduce VISTA3D, Versatile Imaging SegmenTation and Annotation model. The model is trained systematically on 11454 volumes encompassing 127 types of human anatomical structures and various lesions and provides accurate out-of-the-box segmentation. The model's design also achieves state-of-the-art zero-shot interactive segmentation in 3D. The novel model design and training recipe represent a promising step toward developing a versatile medical image foundation model. Code and model weights will be released shortly. The early version of online demo can be tried on https://build.nvidia.com/nvidia/vista-3d.
Automated 3D Segmentation of Kidneys and Tumors in MICCAI KiTS 2023 Challenge
Oct 06, 2023


Abstract:Kidney and Kidney Tumor Segmentation Challenge (KiTS) 2023 offers a platform for researchers to compare their solutions to segmentation from 3D CT. In this work, we describe our submission to the challenge using automated segmentation of Auto3DSeg available in MONAI. Our solution achieves the average dice of 0.835 and surface dice of 0.723, which ranks first and wins the KiTS 2023 challenge.
Aorta Segmentation from 3D CT in MICCAI SEG.A. 2023 Challenge
Oct 06, 2023Abstract:Aorta provides the main blood supply of the body. Screening of aorta with imaging helps for early aortic disease detection and monitoring. In this work, we describe our solution to the Segmentation of the Aorta (SEG.A.231) from 3D CT challenge. We use automated segmentation method Auto3DSeg available in MONAI. Our solution achieves an average Dice score of 0.920 and 95th percentile of the Hausdorff Distance (HD95) of 6.013, which ranks first and wins the SEG.A. 2023 challenge.
Disruptive Autoencoders: Leveraging Low-level features for 3D Medical Image Pre-training
Jul 31, 2023Abstract:Harnessing the power of pre-training on large-scale datasets like ImageNet forms a fundamental building block for the progress of representation learning-driven solutions in computer vision. Medical images are inherently different from natural images as they are acquired in the form of many modalities (CT, MR, PET, Ultrasound etc.) and contain granulated information like tissue, lesion, organs etc. These characteristics of medical images require special attention towards learning features representative of local context. In this work, we focus on designing an effective pre-training framework for 3D radiology images. First, we propose a new masking strategy called local masking where the masking is performed across channel embeddings instead of tokens to improve the learning of local feature representations. We combine this with classical low-level perturbations like adding noise and downsampling to further enable low-level representation learning. To this end, we introduce Disruptive Autoencoders, a pre-training framework that attempts to reconstruct the original image from disruptions created by a combination of local masking and low-level perturbations. Additionally, we also devise a cross-modal contrastive loss (CMCL) to accommodate the pre-training of multiple modalities in a single framework. We curate a large-scale dataset to enable pre-training of 3D medical radiology images (MRI and CT). The proposed pre-training framework is tested across multiple downstream tasks and achieves state-of-the-art performance. Notably, our proposed method tops the public test leaderboard of BTCV multi-organ segmentation challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge