Zhangxing Bian
Likelihood-Separable Diffusion Inference for Multi-Image MRI Super-Resolution
Jan 20, 2026Abstract:Diffusion models are the current state-of-the-art for solving inverse problems in imaging. Their impressive generative capability allows them to approximate sampling from a prior distribution, which alongside a known likelihood function permits posterior sampling without retraining the model. While recent methods have made strides in advancing the accuracy of posterior sampling, the majority focuses on single-image inverse problems. However, for modalities such as magnetic resonance imaging (MRI), it is common to acquire multiple complementary measurements, each low-resolution along a different axis. In this work, we generalize common diffusion-based inverse single-image problem solvers for multi-image super-resolution (MISR) MRI. We show that the DPS likelihood correction allows an exactly-separable gradient decomposition across independently acquired measurements, enabling MISR without constructing a joint operator, modifying the diffusion model, or increasing network function evaluations. We derive MISR versions of DPS, DMAP, DPPS, and diffusion-based PnP/ADMM, and demonstrate substantial gains over SISR across $4\times/8\times/16\times$ anisotropic degradations. Our results achieve state-of-the-art super-resolution of anisotropic MRI volumes and, critically, enable reconstruction of near-isotropic anatomy from routine 2D multi-slice acquisitions, which are otherwise highly degraded in orthogonal views.
Segmenting Thalamic Nuclei: T1 Maps Provide a Reliable and Efficient Solution
Aug 17, 2025Abstract:Accurate thalamic nuclei segmentation is crucial for understanding neurological diseases, brain functions, and guiding clinical interventions. However, the optimal inputs for segmentation remain unclear. This study systematically evaluates multiple MRI contrasts, including MPRAGE and FGATIR sequences, quantitative PD and T1 maps, and multiple T1-weighted images at different inversion times (multi-TI), to determine the most effective inputs. For multi-TI images, we employ a gradient-based saliency analysis with Monte Carlo dropout and propose an Overall Importance Score to select the images contributing most to segmentation. A 3D U-Net is trained on each of these configurations. Results show that T1 maps alone achieve strong quantitative performance and superior qualitative outcomes, while PD maps offer no added value. These findings underscore the value of T1 maps as a reliable and efficient input among the evaluated options, providing valuable guidance for optimizing imaging protocols when thalamic structures are of clinical or research interest.
Brightness-Invariant Tracking Estimation in Tagged MRI
May 23, 2025Abstract:Magnetic resonance (MR) tagging is an imaging technique for noninvasively tracking tissue motion in vivo by creating a visible pattern of magnetization saturation (tags) that deforms with the tissue. Due to longitudinal relaxation and progression to steady-state, the tags and tissue brightnesses change over time, which makes tracking with optical flow methods error-prone. Although Fourier methods can alleviate these problems, they are also sensitive to brightness changes as well as spectral spreading due to motion. To address these problems, we introduce the brightness-invariant tracking estimation (BRITE) technique for tagged MRI. BRITE disentangles the anatomy from the tag pattern in the observed tagged image sequence and simultaneously estimates the Lagrangian motion. The inherent ill-posedness of this problem is addressed by leveraging the expressive power of denoising diffusion probabilistic models to represent the probabilistic distribution of the underlying anatomy and the flexibility of physics-informed neural networks to estimate biologically-plausible motion. A set of tagged MR images of a gel phantom was acquired with various tag periods and imaging flip angles to demonstrate the impact of brightness variations and to validate our method. The results show that BRITE achieves more accurate motion and strain estimates as compared to other state of the art methods, while also being resistant to tag fading.
Unsupervised learning of spatially varying regularization for diffeomorphic image registration
Dec 23, 2024Abstract:Spatially varying regularization accommodates the deformation variations that may be necessary for different anatomical regions during deformable image registration. Historically, optimization-based registration models have harnessed spatially varying regularization to address anatomical subtleties. However, most modern deep learning-based models tend to gravitate towards spatially invariant regularization, wherein a homogenous regularization strength is applied across the entire image, potentially disregarding localized variations. In this paper, we propose a hierarchical probabilistic model that integrates a prior distribution on the deformation regularization strength, enabling the end-to-end learning of a spatially varying deformation regularizer directly from the data. The proposed method is straightforward to implement and easily integrates with various registration network architectures. Additionally, automatic tuning of hyperparameters is achieved through Bayesian optimization, allowing efficient identification of optimal hyperparameters for any given registration task. Comprehensive evaluations on publicly available datasets demonstrate that the proposed method significantly improves registration performance and enhances the interpretability of deep learning-based registration, all while maintaining smooth deformations.
RATNUS: Rapid, Automatic Thalamic Nuclei Segmentation using Multimodal MRI inputs
Sep 10, 2024



Abstract:Accurate segmentation of thalamic nuclei is important for better understanding brain function and improving disease treatment. Traditional segmentation methods often rely on a single T1-weighted image, which has limited contrast in the thalamus. In this work, we introduce RATNUS, which uses synthetic T1-weighted images with many inversion times along with diffusion-derived features to enhance the visibility of nuclei within the thalamus. Using these features, a convolutional neural network is used to segment 13 thalamic nuclei. For comparison with other methods, we introduce a unified nuclei labeling scheme. Our results demonstrate an 87.19% average true positive rate (TPR) against manual labeling. In comparison, FreeSurfer and THOMAS achieve TPRs of 64.25% and 57.64%, respectively, demonstrating the superiority of RATNUS in thalamic nuclei segmentation.
From Registration Uncertainty to Segmentation Uncertainty
Mar 08, 2024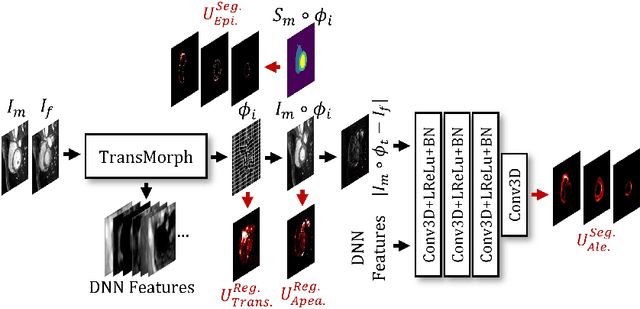
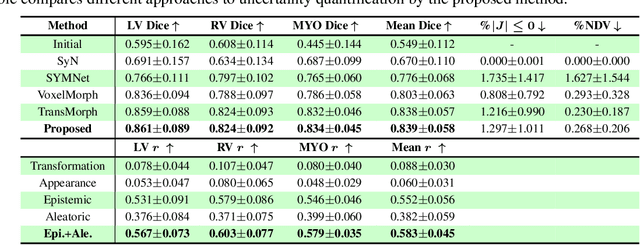
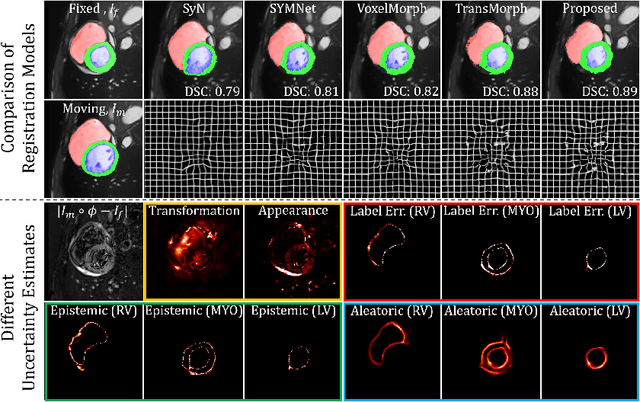
Abstract:Understanding the uncertainty inherent in deep learning-based image registration models has been an ongoing area of research. Existing methods have been developed to quantify both transformation and appearance uncertainties related to the registration process, elucidating areas where the model may exhibit ambiguity regarding the generated deformation. However, our study reveals that neither uncertainty effectively estimates the potential errors when the registration model is used for label propagation. Here, we propose a novel framework to concurrently estimate both the epistemic and aleatoric segmentation uncertainties for image registration. To this end, we implement a compact deep neural network (DNN) designed to transform the appearance discrepancy in the warping into aleatoric segmentation uncertainty by minimizing a negative log-likelihood loss function. Furthermore, we present epistemic segmentation uncertainty within the label propagation process as the entropy of the propagated labels. By introducing segmentation uncertainty along with existing methods for estimating registration uncertainty, we offer vital insights into the potential uncertainties at different stages of image registration. We validated our proposed framework using publicly available datasets, and the results prove that the segmentation uncertainties estimated with the proposed method correlate well with errors in label propagation, all while achieving superior registration performance.
Is Registering Raw Tagged-MR Enough for Strain Estimation in the Era of Deep Learning?
Jan 31, 2024Abstract:Magnetic Resonance Imaging with tagging (tMRI) has long been utilized for quantifying tissue motion and strain during deformation. However, a phenomenon known as tag fading, a gradual decrease in tag visibility over time, often complicates post-processing. The first contribution of this study is to model tag fading by considering the interplay between $T_1$ relaxation and the repeated application of radio frequency (RF) pulses during serial imaging sequences. This is a factor that has been overlooked in prior research on tMRI post-processing. Further, we have observed an emerging trend of utilizing raw tagged MRI within a deep learning-based (DL) registration framework for motion estimation. In this work, we evaluate and analyze the impact of commonly used image similarity objectives in training DL registrations on raw tMRI. This is then compared with the Harmonic Phase-based approach, a traditional approach which is claimed to be robust to tag fading. Our findings, derived from both simulated images and an actual phantom scan, reveal the limitations of various similarity losses in raw tMRI and emphasize caution in registration tasks where image intensity changes over time.
Efficient Annotation for Medical Image Analysis: A One-Pass Selective Annotation Approach
Sep 15, 2023



Abstract:Annotating biomedical images for supervised learning is a complex and labor-intensive task due to data diversity and its intricate nature. In this paper, we propose an innovative method, the efficient one-pass selective annotation (EPOSA), that significantly reduces the annotation burden while maintaining robust model performance. Our approach employs a variational autoencoder (VAE) to extract salient features from unannotated images, which are subsequently clustered using the DBSCAN algorithm. This process groups similar images together, forming distinct clusters. We then use a two-stage sample selection algorithm, called representative selection (RepSel), to form a selected dataset. The first stage is a Markov Chain Monte Carlo (MCMC) sampling technique to select representative samples from each cluster for annotations. This selection process is the second stage, which is guided by the principle of maximizing intra-cluster mutual information and minimizing inter-cluster mutual information. This ensures a diverse set of features for model training and minimizes outlier inclusion. The selected samples are used to train a VGG-16 network for image classification. Experimental results on the Med-MNIST dataset demonstrate that our proposed EPOSA outperforms random selection and other state-of-the-art methods under the same annotation budget, presenting a promising direction for efficient and effective annotation in medical image analysis.
MomentaMorph: Unsupervised Spatial-Temporal Registration with Momenta, Shooting, and Correction
Aug 05, 2023



Abstract:Tagged magnetic resonance imaging (tMRI) has been employed for decades to measure the motion of tissue undergoing deformation. However, registration-based motion estimation from tMRI is difficult due to the periodic patterns in these images, particularly when the motion is large. With a larger motion the registration approach gets trapped in a local optima, leading to motion estimation errors. We introduce a novel "momenta, shooting, and correction" framework for Lagrangian motion estimation in the presence of repetitive patterns and large motion. This framework, grounded in Lie algebra and Lie group principles, accumulates momenta in the tangent vector space and employs exponential mapping in the diffeomorphic space for rapid approximation towards true optima, circumventing local optima. A subsequent correction step ensures convergence to true optima. The results on a 2D synthetic dataset and a real 3D tMRI dataset demonstrate our method's efficiency in estimating accurate, dense, and diffeomorphic 2D/3D motion fields amidst large motion and repetitive patterns.
A Survey on Deep Learning in Medical Image Registration: New Technologies, Uncertainty, Evaluation Metrics, and Beyond
Jul 28, 2023



Abstract:Over the past decade, deep learning technologies have greatly advanced the field of medical image registration. The initial developments, such as ResNet-based and U-Net-based networks, laid the groundwork for deep learning-driven image registration. Subsequent progress has been made in various aspects of deep learning-based registration, including similarity measures, deformation regularizations, and uncertainty estimation. These advancements have not only enriched the field of deformable image registration but have also facilitated its application in a wide range of tasks, including atlas construction, multi-atlas segmentation, motion estimation, and 2D-3D registration. In this paper, we present a comprehensive overview of the most recent advancements in deep learning-based image registration. We begin with a concise introduction to the core concepts of deep learning-based image registration. Then, we delve into innovative network architectures, loss functions specific to registration, and methods for estimating registration uncertainty. Additionally, this paper explores appropriate evaluation metrics for assessing the performance of deep learning models in registration tasks. Finally, we highlight the practical applications of these novel techniques in medical imaging and discuss the future prospects of deep learning-based image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge